Organization of the pre-autophagosomal structure responsible for autophagosome formation
- PMID: 18287526
- PMCID: PMC2366851
- DOI: 10.1091/mbc.e07-10-1048
Organization of the pre-autophagosomal structure responsible for autophagosome formation
Abstract
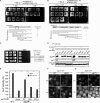
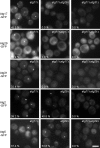
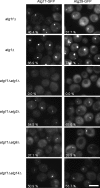
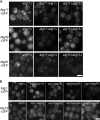
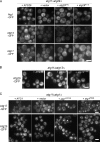
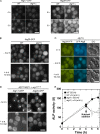
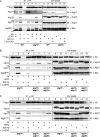
Autophagy induced by nutrient depletion is involved in survival during starvation conditions. In addition to starvation-induced autophagy, the yeast Saccharomyces cerevisiae also has a constitutive autophagy-like system, the Cvt pathway. Among 31 autophagy-related (Atg) proteins, the function of Atg17, Atg29, and Atg31 is required specifically for autophagy. In this study, we investigated the role of autophagy-specific (i.e., non-Cvt) proteins under autophagy-inducing conditions. For this purpose, we used atg11Delta cells in which the Cvt pathway is abrogated. The autophagy-unique proteins are required for the localization of Atg proteins to the pre-autophagosomal structure (PAS), the putative site for autophagosome formation, under starvation condition. It is likely that these Atg proteins function as a ternary complex, because Atg29 and Atg31 bind to Atg17. The Atg1 kinase complex (Atg1-Atg13) is also essential for recruitment of Atg proteins to the PAS. The assembly of Atg proteins to the PAS is observed only under autophagy-inducing conditions, indicating that this structure is specifically involved in autophagosome formation. Our results suggest that Atg1 complex and the autophagy-unique Atg proteins cooperatively organize the PAS in response to starvation signals.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

