FBXO25-associated nuclear domains: a novel subnuclear structure
- PMID: 18287534
- PMCID: PMC2366848
- DOI: 10.1091/mbc.e07-08-0815
FBXO25-associated nuclear domains: a novel subnuclear structure
Abstract
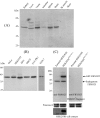
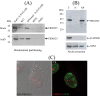
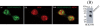
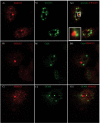
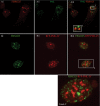
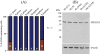
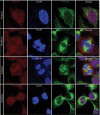
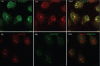
Skp1, Cul1, Rbx1, and the FBXO25 protein form a functional ubiquitin ligase complex. Here, we investigate the cellular distribution of FBXO25 and its colocalization with some nuclear proteins by using immunochemical and biochemical approaches. FBXO25 was monitored with affinity-purified antibodies raised against the recombinant fragment spanning residues 2-62 of the FBXO25 sequence. FBXO25 protein was expressed in all mouse tissues tested except striated muscle, as indicated by immunoblot analysis. Confocal analysis revealed that the endogenous FBXO25 was partially concentrated in a novel dot-like nuclear domain that is distinct from clastosomes and other well-characterized structures. These nuclear compartments contain a high concentration of ubiquitin conjugates and at least two other components of the ubiquitin-proteasome system: 20S proteasome and Skp1. We propose to name these compartments FBXO25-associated nuclear domains. Interestingly, inhibition of transcription by actinomycin D or heat-shock treatment drastically affected the nuclear organization of FBXO25-containing structures, indicating that they are dynamic compartments influenced by the transcriptional activity of the cell. Also, we present evidences that an FBXO25-dependent ubiquitin ligase activity prevents aggregation of recombinant polyglutamine-containing huntingtin protein in the nucleus of human embryonic kidney 293 cells, suggesting that this protein can be a target for the nuclear FBXO25 mediated ubiquitination.
Figures




Similar articles
-
Identification of FBXO25-interacting proteins using an integrated proteomics approach.Proteomics. 2010 Aug;10(15):2746-57. doi: 10.1002/pmic.200900419. Proteomics. 2010. PMID: 20473970 Free PMC article.
-
A novel Fbxo25 acts as an E3 ligase for destructing cardiac specific transcription factors.Biochem Biophys Res Commun. 2011 Jul 1;410(2):183-8. doi: 10.1016/j.bbrc.2011.05.011. Epub 2011 May 7. Biochem Biophys Res Commun. 2011. PMID: 21596019
-
FBXO25, an F-box protein homologue of atrogin-1, is not induced in atrophying muscle.Biochim Biophys Acta. 2006 Jun;1760(6):966-72. doi: 10.1016/j.bbagen.2006.03.020. Epub 2006 Apr 4. Biochim Biophys Acta. 2006. PMID: 16714087
-
Nuclear speckles: a model for nuclear organelles.Nat Rev Mol Cell Biol. 2003 Aug;4(8):605-12. doi: 10.1038/nrm1172. Nat Rev Mol Cell Biol. 2003. PMID: 12923522 Review.
-
Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases.Cell Res. 2013 May;23(5):599-619. doi: 10.1038/cr.2013.44. Epub 2013 Mar 26. Cell Res. 2013. PMID: 23528706 Free PMC article. Review.
Cited by
-
Lipid microdomains in cell nucleus.Mol Biol Cell. 2008 Dec;19(12):5289-95. doi: 10.1091/mbc.e08-05-0517. Epub 2008 Oct 15. Mol Biol Cell. 2008. PMID: 18923143 Free PMC article.
-
FBXO25 promotes cell proliferation, invasion, and migration of NSCLC.Tumour Biol. 2016 Oct;37(10):14311-14319. doi: 10.1007/s13277-016-5298-1. Epub 2016 Sep 5. Tumour Biol. 2016. PMID: 27596142
-
The F-box protein FBXO25 promotes the proteasome-dependent degradation of ELK-1 protein.J Biol Chem. 2013 Sep 27;288(39):28152-62. doi: 10.1074/jbc.M113.504308. Epub 2013 Aug 12. J Biol Chem. 2013. PMID: 23940030 Free PMC article.
-
An Integrated Approach to Explore Composition and Dynamics of Cholesterol-rich Membrane Microdomains in Sexual Stages of Malaria Parasite.Mol Cell Proteomics. 2017 Oct;16(10):1801-1814. doi: 10.1074/mcp.M117.067041. Epub 2017 Aug 10. Mol Cell Proteomics. 2017. PMID: 28798222 Free PMC article.
-
Role of ERLINs in the Control of Cell Fate through Lipid Rafts.Cells. 2021 Sep 13;10(9):2408. doi: 10.3390/cells10092408. Cells. 2021. PMID: 34572057 Free PMC article. Review.
References
-
- Ang X. L., Harper J. W. Interwoven ubiquitination oscillators and control of cell cycle transitions. Oncogene. 2005;17:2860–2870. - PubMed
-
- Ardley H. C., Robinson P. A. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. - PubMed
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D, Seidman J. G., Smith J. A., Struhl K. Short Protocols in Molecular Biology. 3rd ed. New York: John Wiley & Sons; 1997.
-
- Carmo-Fonseca M., Mendes-Soares L., Campos I. To be or not to be in the nucleolus. Nat. Cell Biol. 2000;2:E107–E112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

