GRIM-19 is essential for maintenance of mitochondrial membrane potential
- PMID: 18287540
- PMCID: PMC2366854
- DOI: 10.1091/mbc.e07-07-0683
GRIM-19 is essential for maintenance of mitochondrial membrane potential
Abstract
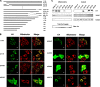
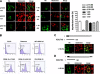
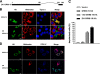
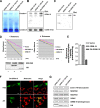
GRIM-19 was found to copurify with complex I of mitochondrial respiratory chain and subsequently was demonstrated to be involved in complex I assembly and activity. To further understand its function in complex I, we dissected its functional domains by generating a number of deletion, truncation, and point mutants. The mitochondrial localization sequences were located at the N-terminus. Strikingly, deletion of residues 70-80, 90-100, or the whole C-terminal region (70-144) led to a loss of mitochondrial transmembrane potential (DeltaPsim). However, similar deletions of another two complex I subunits, NDUFA9 and NDUFS3, did not show such effect. We also found that deletion of the last 10 residues affected GRIM-19's ability to be assembled to complex I. We constructed a dominant-negative mutant containing the N-terminal 60 and the last C-terminal 10 residues, which could be assembled into complex I, but failed to maintain normal DeltaPsim. Cells overexpressing this mutant did not spontaneously undergo cell death, but were sensitized to apoptosis induced by cell death agents. Our results demonstrate that GRIM-19 is required for electron transfer activity of complex I, and disruption of DeltaPsim by GRIM-19 mutants enhances the cells' sensitivity to apoptotic stimuli.
Figures



References
-
- Angell J. E., Lindner D. J., Shapiro P. S., Hofmann E. R., Kalvakolanu D. V. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J. Biol. Chem. 2000;275:33416–33426. - PubMed
-
- Appleby R. D., Porteous W. K., Hughes G., James A. M., Shannon D., Wei Y. H., Murphy M. P. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 1999;262:108–116. - PubMed
-
- Carroll J., Fearnley I. M., Shannon R. J., Hirst J., Walker J. E. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell Proteomics. 2003;2:117–126. - PubMed
-
- Carroll J., Fearnley I. M., Skehel J. M., Runswick M. J., Shannon R. J., Hirst J., Walker J. E. The post-translational modifications of the nuclear encoded subunits of complex I from bovine heart mitochondria. Mol. Cell. Proteomics. 2005;4:693–699. - PubMed
-
- Carroll J., Fearnley I. M., Skehel J. M., Shannon R. J., Hirst J., Walker J. E. Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 2006;281:32724–32727. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

