Structural lability in stem-loop 1 drives a 5' UTR-3' UTR interaction in coronavirus replication
- PMID: 18289557
- PMCID: PMC2652258
- DOI: 10.1016/j.jmb.2008.01.068
Structural lability in stem-loop 1 drives a 5' UTR-3' UTR interaction in coronavirus replication
Abstract
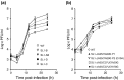
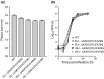
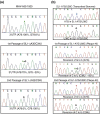
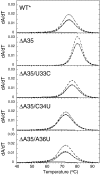
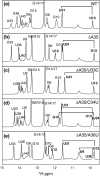
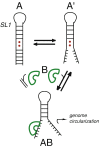
The leader RNA of the 5' untranslated region (UTR) of coronaviral genomes contains two stem-loop structures denoted SL1 and SL2. Herein, we show that SL1 is functionally and structurally bipartite. While the upper region of SL1 is required to be paired, we observe strong genetic selection against viruses that contain a deletion of A35, an extrahelical nucleotide that destabilizes SL1, in favor of genomes that contain a diverse panel of destabilizing second-site mutations, due to introduction of a noncanonical base pair near A35. Viruses containing destabilizing SL1-DeltaA35 mutations also contain one of two specific mutations in the 3' UTR. Thermal denaturation and imino proton solvent exchange experiments reveal that the lower half of SL1 is unstable and that second-site SL1-DeltaA35 substitutions are characterized by one or more features of the wild-type SL1. We propose a "dynamic SL1" model, in which the base of SL1 has an optimized lability required to mediate a physical interaction between the 5' UTR and the 3' UTR that stimulates subgenomic RNA synthesis. Although not conserved at the nucleotide sequence level, these general structural characteristics of SL1 appear to be conserved in other coronaviral genomes.
Figures
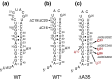


Similar articles
-
Mouse hepatitis virus stem-loop 4 functions as a spacer element required to drive subgenomic RNA synthesis.J Virol. 2011 Sep;85(17):9199-209. doi: 10.1128/JVI.05092-11. Epub 2011 Jun 29. J Virol. 2011. PMID: 21715502 Free PMC article.
-
A U-turn motif-containing stem-loop in the coronavirus 5' untranslated region plays a functional role in replication.RNA. 2007 May;13(5):763-80. doi: 10.1261/rna.261807. Epub 2007 Mar 12. RNA. 2007. PMID: 17353353 Free PMC article.
-
The 3' end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5' end region.J Gen Virol. 2006 Oct;87(Pt 10):3013-3022. doi: 10.1099/vir.0.82059-0. J Gen Virol. 2006. PMID: 16963760
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
-
End-to-end communication in the modulation of translation by mammalian RNA viruses.Virus Res. 2006 Jul;119(1):43-51. doi: 10.1016/j.virusres.2005.10.012. Epub 2005 Nov 22. Virus Res. 2006. PMID: 16307817 Free PMC article. Review.
Cited by
-
Dissection of amino-terminal functional domains of murine coronavirus nonstructural protein 3.J Virol. 2015 Jun;89(11):6033-47. doi: 10.1128/JVI.00197-15. Epub 2015 Mar 25. J Virol. 2015. PMID: 25810552 Free PMC article.
-
SHAPE analysis of the RNA secondary structure of the Mouse Hepatitis Virus 5' untranslated region and N-terminal nsp1 coding sequences.Virology. 2015 Jan 15;475:15-27. doi: 10.1016/j.virol.2014.11.001. Epub 2014 Nov 21. Virology. 2015. PMID: 25462342 Free PMC article.
-
Group-specific structural features of the 5'-proximal sequences of coronavirus genomic RNAs.Virology. 2010 May 25;401(1):29-41. doi: 10.1016/j.virol.2010.02.007. Epub 2010 Mar 4. Virology. 2010. PMID: 20202661 Free PMC article.
-
Lessons Learned and Yet-to-Be Learned on the Importance of RNA Structure in SARS-CoV-2 Replication.Microbiol Mol Biol Rev. 2022 Sep 21;86(3):e0005721. doi: 10.1128/mmbr.00057-21. Epub 2022 Jul 7. Microbiol Mol Biol Rev. 2022. PMID: 35862724 Free PMC article. Review.
-
Interaction of coronavirus nucleocapsid protein with the 5'- and 3'-ends of the coronavirus genome is involved in genome circularization and negative-strand RNA synthesis.FEBS J. 2019 Aug;286(16):3222-3239. doi: 10.1111/febs.14863. Epub 2019 May 8. FEBS J. 2019. PMID: 31034708 Free PMC article.
References
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
-
- Weiss S.R., Leibowitz J.L. Pathogenesis of murine coronavirus infections. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; Washington, DC: 2007. pp. 259–278.
-
- Breedenbeek P.J., Pachuk C.J., Noten A.F.H., Charité J., Luyjtes W., Weiss S.R., Spaan W.J.M. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

