Computational identification of the normal and perturbed genetic networks involved in myeloid differentiation and acute promyelocytic leukemia
- PMID: 18291030
- PMCID: PMC2374711
- DOI: 10.1186/gb-2008-9-2-r38
Computational identification of the normal and perturbed genetic networks involved in myeloid differentiation and acute promyelocytic leukemia
Abstract
Background: Acute myeloid leukemia (AML) comprises a group of diseases characterized by the abnormal development of malignant myeloid cells. Recent studies have demonstrated an important role for aberrant transcriptional regulation in AML pathophysiology. Although several transcription factors (TFs) involved in myeloid development and leukemia have been studied extensively and independently, how these TFs coordinate with others and how their dysregulation perturbs the genetic circuitry underlying myeloid differentiation is not yet known. We propose an integrated approach for mammalian genetic network construction by combining the analysis of gene expression profiling data and the identification of TF binding sites.
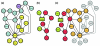
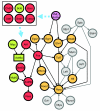
Results: We utilized our approach to construct the genetic circuitries operating in normal myeloid differentiation versus acute promyelocytic leukemia (APL), a subtype of AML. In the normal and disease networks, we found that multiple transcriptional regulatory cascades converge on the TFs Rora and Rxra, respectively. Furthermore, the TFs dysregulated in APL participate in a common regulatory pathway and may perturb the normal network through Fos. Finally, a model of APL pathogenesis is proposed in which the chimeric TF PML-RARalpha activates the dysregulation in APL through six mediator TFs.
Conclusion: This report demonstrates the utility of our approach to construct mammalian genetic networks, and to obtain new insights regarding regulatory circuitries operating in complex diseases in humans.
Figures



References
-
- Helbling D, Mueller BU, Timchenko NA, Hagemeijer A, Jotterand M, Meyer-Monard S, Lister A, Rowley JD, Huegli B, Fey MF, Pabst T. The leukemic fusion gene AML1-MDS1-EVI1 suppresses CEBPA in acute myeloid leukemia by activation of Calreticulin. Proc Natl Acad Sci USA. 2004;101:13312–13317. doi: 10.1073/pnas.0404731101. - DOI - PMC - PubMed
-
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. - PubMed
-
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

