New applications of the Comet assay: Comet-FISH and transcription-coupled DNA repair
- PMID: 18291710
- PMCID: PMC2667151
- DOI: 10.1016/j.mrrev.2007.12.003
New applications of the Comet assay: Comet-FISH and transcription-coupled DNA repair
Abstract
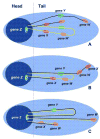
Transcription-coupled repair (TCR) is a pathway dedicated to the removal of damage from the template strands of actively transcribed genes. Although the detailed mechanism of TCR is not yet understood, it is believed to be triggered when a translocating RNA polymerase is arrested at a lesion or unusual structure in the DNA. Conventional assays for TCR require high doses of DNA damage for the statistical analysis of repair in the individual strands of DNA sequences ranging in size from a few hundred bases to 30kb. The single cell gel electrophoresis (Comet) assay allows detection of single- or double-strand breaks at a 10-100-fold higher level of resolution. Fluorescence in situ hybridization (FISH) combined with the Comet assay (Comet-FISH) affords a heightened level of sensitivity for the assessment of repair in defined DNA sequences of cells treated with physiologically relevant doses of genotoxins. This approach also reveals localized susceptibility to chromosomal breakage in cells from individuals with hypersensitivity to radiation or chemotherapy. Several groups have reported preferential repair in transcriptionally active genes or chromosomal domains using Comet-FISH. The prevailing interpretation of the behavior of DNA in the Comet assay assumes that the DNA is arranged in loops and matrix-attachment sites; that supercoiled, undamaged loops are contained within the nuclear matrix and appear in Comet "heads", and that Comet "tails" consist of relaxed DNA loops containing one or more breaks. According to this model, localization of FISH probes in Comet heads signifies that loops containing the targeted sequences are free of damage. This implies that preferential repair as detected by Comet-FISH might encompass large chromosomal domains containing both transcribed and non-transcribed sequences. We review the existing evidence and discuss the implications in relation to current models for the molecular mechanism of TCR.
Figures

Similar articles
-
Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells.Nucleic Acids Res. 2013 Sep;41(16):7700-12. doi: 10.1093/nar/gkt524. Epub 2013 Jun 17. Nucleic Acids Res. 2013. PMID: 23775797 Free PMC article.
-
Analysis of DNA damage and repair by comet fluorescence in situ hybridization (Comet-FISH).Methods Mol Biol. 2014;1094:39-48. doi: 10.1007/978-1-62703-706-8_4. Methods Mol Biol. 2014. PMID: 24162978
-
Detection of DNA damage by comet fluorescence in situ hybridization.Methods Mol Biol. 2012;920:91-100. doi: 10.1007/978-1-61779-998-3_7. Methods Mol Biol. 2012. PMID: 22941598
-
Use of Comet-FISH in the study of DNA damage and repair: review.Mutat Res. 2009 Jan-Feb;681(1):33-43. doi: 10.1016/j.mrrev.2008.01.006. Epub 2008 Jan 31. Mutat Res. 2009. PMID: 18304859 Review.
-
Increasing the resolution of the comet assay using fluorescent in situ hybridization--a review.Mutagenesis. 2009 Sep;24(5):383-9. doi: 10.1093/mutage/gep021. Epub 2009 Jun 17. Mutagenesis. 2009. PMID: 19535362 Review.
Cited by
-
Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells.Nucleic Acids Res. 2013 Sep;41(16):7700-12. doi: 10.1093/nar/gkt524. Epub 2013 Jun 17. Nucleic Acids Res. 2013. PMID: 23775797 Free PMC article.
-
New developments in comet-FISH.Mutagenesis. 2015 Jan;30(1):5-9. doi: 10.1093/mutage/geu036. Mutagenesis. 2015. PMID: 25527722 Free PMC article.
-
UVSSA and USP7, a new couple in transcription-coupled DNA repair.Chromosoma. 2013 Aug;122(4):275-84. doi: 10.1007/s00412-013-0420-2. Epub 2013 Jun 13. Chromosoma. 2013. PMID: 23760561 Free PMC article. Review.
-
True lies: the double life of the nucleotide excision repair factors in transcription and DNA repair.J Nucleic Acids. 2010 Jul 25;2010:616342. doi: 10.4061/2010/616342. J Nucleic Acids. 2010. PMID: 20725631 Free PMC article.
-
Strand-specific PCR of UV radiation-damaged genomic DNA revealed an essential role of DNA-PKcs in the transcription-coupled repair.BMC Biochem. 2011 Jan 8;12:2. doi: 10.1186/1471-2091-12-2. BMC Biochem. 2011. PMID: 21214942 Free PMC article.
References
-
- Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington DC: 2006.
-
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient that in the genome overall. Cell. 1985;40:359–369. - PubMed
-
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. - PubMed
-
- Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. - PubMed
-
- Leadon SA, Lawrence DA. Strand-selective repair of DNA damage in the yeast gal7 gene requires RNA polymerase-II. J Biol Chem. 1992;267:23175–23182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

