Tumor necrosis factor (TNF)-alpha persistently activates nuclear factor-kappaB signaling through the type 2 TNF receptor in chromaffin cells: implications for long-term regulation of neuropeptide gene expression in inflammation
- PMID: 18292192
- PMCID: PMC2408812
- DOI: 10.1210/en.2007-1192
Tumor necrosis factor (TNF)-alpha persistently activates nuclear factor-kappaB signaling through the type 2 TNF receptor in chromaffin cells: implications for long-term regulation of neuropeptide gene expression in inflammation
Abstract
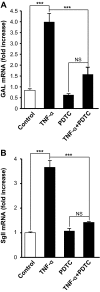
Chromaffin cells of the adrenal medulla elaborate and secrete catecholamines and neuropeptides for hormonal and paracrine signaling in stress and during inflammation. We have recently documented the action of the cytokine TNF-alpha on neuropeptide secretion and biosynthesis in isolated bovine chromaffin cells. Here, we demonstrate that the type 2 TNF-alpha receptor (TNF-R2) mediates TNF-alpha signaling in chromaffin cells via activation of nuclear factor (NF)-kappaB. Microarray and suppression subtractive hybridization have been used to identify TNF-alpha target genes in addition to those encoding the neuropeptides galanin, vasoactive intestinal polypeptide, and secretogranin II in chromaffin cells. TNF-alpha, acting through the TNF-R2, causes an early up-regulation of NF-kappaB, long-lasting induction of the NF-kappaB target gene inhibitor kappaB (IkappaB), and persistent stimulation of other NF-kappaB-associated genes including mitogen-inducible gene-6 (MIG-6), which acts as an IkappaB signaling antagonist, and butyrate-induced transcript 1. Consistent with long-term activation of the NF-kappaB signaling pathway, delayed induction of neuropeptide gene transcription by TNF-alpha in chromaffin cells is blocked by an antagonist of NF-kappaB signaling. TNF-alpha-dependent signaling in neuroendocrine cells thus leads to a unique, persistent mode of NF-kappaB activation that features long-lasting transcription of both IkappaB and MIG-6, which may play a role in the long-lasting effects of TNF-alpha in regulating neuropeptide output from the adrenal, a potentially important feedback station for modulating long-term cytokine effects in inflammation.
Figures
References
-
- Wrona D 2006 Neural-immune interactions: an integrative view of the bidirectional relationship between the brain and immune systems. J Neuroimmunol 172:38–58 - PubMed
-
- Andreis PG, Tortorella C, Ziolkowska A, Spinazzi R, Malendowicz LK, Neri G, Nussdorfer GG 2007 Evidence for a paracrine role of endogenous adrenomedullary galanin in the regulation of glucocorticoid secretion in the rat adrenal gland. Int J Mol Med 19:511–515 - PubMed
-
- Ait-Ali D, Turquier V, Grumolato L, Yon L, Jourdain M, Alexandre D, Eiden LE, Vaudry H, Anouar Y 2004 The proinflammatory cytokines tumor necrosis factor-α and interleukin-1 stimulate neuropeptide gene transcription and secretion in adrenochromaffin cells via activation of extracellularly regulated kinase 1/2 and p38 protein kinases, and activator protein-1 transcription factors. Mol Endocrinol 18:1721–1739 - PubMed
-
- Bornstein SR, Haidan A, Ehrhart-Bornstein M 1996 Cellular communication in the neuro-adrenocortical axis: role of vasoactive intestinal polypeptide (VIP). Endocr Res 22:819–829 - PubMed
-
- Kruppa G, Thoma B, Machleidt T, Wiegmann K, Kronke M 1992 Inhibition of tumor necrosis factor (TNF)-mediated NF-κB activation by selective blockade of the human 55-kDa TNF receptor. J Immunol 148:3152–3157 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

