A natural polymorphism in peroxisome proliferator-activated receptor-alpha hinge region attenuates transcription due to defective release of nuclear receptor corepressor from chromatin
- PMID: 18292238
- PMCID: PMC5419521
- DOI: 10.1210/me.2007-0547
A natural polymorphism in peroxisome proliferator-activated receptor-alpha hinge region attenuates transcription due to defective release of nuclear receptor corepressor from chromatin
Abstract
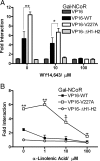
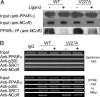
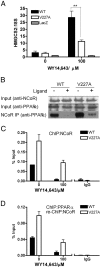
Peroxisome proliferator-activated receptor-alpha (PPARalpha) is a central regulator of lipid metabolism. Fibrate drugs act on PPARalpha to modulate dyslipidemias. A natural variant (V227A) affecting the PPARalpha hinge region was associated with perturbations in blood lipid levels in Asian populations. In this study, we investigated the functional significance of the V227A substitution. The variant significantly attenuated PPARalpha-mediated transactivation of the cytochrome P450 4A6 and mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase (HMGCS2) genes in the presence of fibrate ligands. Screening of a panel of PPARalpha coregulators revealed that V227A enhanced recruitment of the nuclear corepressor NCoR. Transactivation activity of V227A could be restored by silencing NCoR or by inhibition of its histone deacetylase activity. Deletion studies indicated that PPARalpha interacted with NCoR receptor-interacting domain 1 (ID1) but not ID2 or ID3. These interactions were dependent on the intact consensus nonapeptide nuclear receptor interaction motif in NCoR ID1 and were enhanced by the adjacent 24 N-terminal residues. Novel corepressor interaction determinants involving PPARalpha helices 1 and 2 were identified. In hepatic cells, the V227A substitution stabilized PPARalpha/NCoR interactions and caused defective release of NCoR in the presence of agonists on the HMGCS2 promoter. These results provide the first indication that defective function of a natural PPARalpha variant was due, at least partially, to increased corepressor binding. Our data suggest that the PPARalpha/NCoR interaction is physiologically relevant and can produce a discernable phenotype when the magnitude of the interaction is altered by a naturally occurring variation.
Figures







Similar articles
-
Nuclear receptor corepressor-dependent repression of peroxisome-proliferator-activated receptor delta-mediated transactivation.Biochem J. 2002 Apr 1;363(Pt 1):157-65. doi: 10.1042/0264-6021:3630157. Biochem J. 2002. PMID: 11903058 Free PMC article.
-
Identification of nuclear receptor corepressor as a peroxisome proliferator-activated receptor alpha interacting protein.J Biol Chem. 1999 May 28;274(22):15901-7. doi: 10.1074/jbc.274.22.15901. J Biol Chem. 1999. PMID: 10336495
-
Function of multiple Lis-Homology domain/WD-40 repeat-containing proteins in feed-forward transcriptional repression by silencing mediator for retinoic and thyroid receptor/nuclear receptor corepressor complexes.Mol Endocrinol. 2008 May;22(5):1093-104. doi: 10.1210/me.2007-0396. Epub 2008 Jan 17. Mol Endocrinol. 2008. PMID: 18202150 Free PMC article.
-
A novel mechanism of thyroid hormone-dependent negative regulation by thyroid hormone receptor, nuclear receptor corepressor (NCoR), and GAGA-binding factor on the rat cD44 promoter.J Biol Chem. 2005 Apr 15;280(15):14545-55. doi: 10.1074/jbc.M411517200. Epub 2005 Jan 27. J Biol Chem. 2005. PMID: 15701601
-
PPARα and NCOR/SMRT corepressor network in liver metabolic regulation.FASEB J. 2020 Jul;34(7):8796-8809. doi: 10.1096/fj.202000055RR. Epub 2020 May 12. FASEB J. 2020. PMID: 32396271 Review.
Cited by
-
General molecular biology and architecture of nuclear receptors.Curr Top Med Chem. 2012;12(6):486-504. doi: 10.2174/156802612799436641. Curr Top Med Chem. 2012. PMID: 22242852 Free PMC article. Review.
-
SUMOylation of human peroxisome proliferator-activated receptor alpha inhibits its trans-activity through the recruitment of the nuclear corepressor NCoR.J Biol Chem. 2010 Feb 26;285(9):5983-92. doi: 10.1074/jbc.M109.078311. Epub 2009 Dec 2. J Biol Chem. 2010. PMID: 19955185 Free PMC article.
-
The Influence of the Differentiation of Genes Encoding Peroxisome Proliferator-Activated Receptors and Their Coactivators on Nutrient and Energy Metabolism.Nutrients. 2022 Dec 18;14(24):5378. doi: 10.3390/nu14245378. Nutrients. 2022. PMID: 36558537 Free PMC article.
-
PPARɑ variant V227A reduces plasma triglycerides through enhanced lipoprotein lipolysis.J Lipid Res. 2025 May;66(5):100806. doi: 10.1016/j.jlr.2025.100806. Epub 2025 Apr 15. J Lipid Res. 2025. PMID: 40245984 Free PMC article.
-
Impact of PPAR-Alpha Polymorphisms-The Case of Metabolic Disorders and Atherosclerosis.Int J Mol Sci. 2019 Sep 6;20(18):4378. doi: 10.3390/ijms20184378. Int J Mol Sci. 2019. PMID: 31489930 Free PMC article. Review.
References
-
- Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O’Rahilly S, Palmer CN, Plutzky J, Reddy JK, Spiegelman BM, Staels B, Wahli W 2006. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev 58:726–741 - PubMed
-
- Desvergne B, Wahli W 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev 20:649–688 - PubMed
-
- Germain P, Staels B, Dacquet C, Spedding M, Laudet V 2006. Overview of nomenclature of nuclear receptors. Pharmacol Rev 58:685–704 - PubMed
-
- Krey G, Keller H, Mahfoudi A, Medin J, Ozato K, Dreyer C, Wahli W 1993. Xenopus peroxisome proliferator activated receptors: genomic organization, response element recognition, heterodimer formation with retinoid X receptor and activation by fatty acids. J Steroid Biochem Mol Biol 47:65–73 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

