Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers
- PMID: 18294333
- PMCID: PMC2432489
- DOI: 10.1111/j.1365-2125.2008.03104.x
Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers
Abstract
What is already known about this subject: The only existing study of CYP2C19*17-associated alterations in drug pharmacokinetics was retrospective and compared probe drug metabolic ratios. The CYP2C19*17 allele had been associated with a two- and fourfold decrease in omeprazole and S/R-mephenytoin metabolic ratios.
What this study adds: This study characterized the single-dose pharmacokinetics of omeprazole, along with the 5-hydroxy and sulphone metabolites, in CYP2C19*17/*17 and CYP2C19*1/*1 subjects. The observed differences in omeprazole AUC(infinity) suggest that the CYP2C19*17 allele is an important explanatory factor behind individual cases of therapeutic failure. AIMS To investigate the influence of the CYP2C19*17 allele on the pharmacokinetics of omeprazole, a commonly used CYP2C19 probe drug, in healthy volunteers.
Methods: In a single-dose pharmacokinetic study, 17 healthy White volunteers genotyped as either CYP2C19*17/*17 or CYP2C19*1/*1 received an oral dose of 40 mg of omeprazole. Plasma was sampled for up to 10 h postdose, followed by quantification of omeprazole, 5-hydroxy omeprazole and omeprazole sulphone by high-performance liquid chromatography.
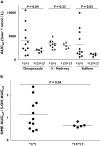
Results: The mean omeprazole AUC(infinity) of 1973 h nmol l(-1) in CYP2C19*17/*17 subjects was 2.1-fold lower [95% confidence interval (CI) 1.1, 3.3] than in CYP2C19*1/*1 subjects (4151 h nmol l(-1), P = 0.04). A similar trend was observed for the sulphone metabolite with the CYP2C19*17/*17 group having a mean AUC(infinity) of 1083 h nmol l(-1), 3.1-fold lower (95% CI 1.2, 5.5) than the CYP2C19*1/*1 group (3343 h nmol l(-1), P = 0.03). A pronounced correlation (r(2) = 0.95, P < 0.0001) was seen in the intraindividual omeprazole AUC(infinity) and omeprazole sulphone AUC(infinity) values.
Conclusions: The pharmacokinetics of omeprazole and omeprazole sulphone differ significantly between homozygous CYP2C19*17 and CYP2C19*1 subjects. For clinically important drugs that are metabolized predominantly by CYP2C19, the CYP2C19*17 allele might be associated with subtherapeutic drug exposure.
Figures

References
-
- Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41:913–58. - PubMed
-
- Bertilsson L, Henthorn TK, Sanz E, Tybring G, Sawe J, Villen T. Importance of genetic factors in the regulation of diazepam metabolism: relationship to S-mephenytoin, but not debrisoquin, hydroxylation phenotype. Clin Pharmacol Ther. 1989;45:348–55. - PubMed
-
- Qin XP, Xie HG, Wang W, He N, Huang SL, Xu ZH, Ou-Yang DS, Wang YJ, Zhou HH. Effect of the gene dosage of CYP2C19 on diazepam metabolism in Chinese subjects. Clin Pharmacol Ther. 1999;66:642–6. - PubMed
-
- Sindrup SH, Brosen K, Hansen MG, Aaes-Jorgensen T, Overo KF, Gram LF. Pharmacokinetics of citalopram in relation to the sparteine and the mephenytoin oxidation polymorphisms. Ther Drug Monit. 1993;15:11–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

