Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans
- PMID: 18294377
- PMCID: PMC2374700
- DOI: 10.1186/gb-2008-9-2-r41
Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans
Abstract
Background: Genome variability can have a profound influence on the virulence of pathogenic microbes. The availability of genome sequences for two strains of the AIDS-associated fungal pathogen Cryptococcus neoformans presented an opportunity to use comparative genome hybridization (CGH) to examine genome variability between strains of different mating type, molecular subtype, and ploidy.
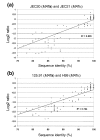
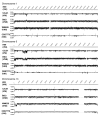
Results: Initially, CGH was used to compare the approximately 100 kilobase MATa and MATalpha mating-type regions in serotype A and D strains to establish the relationship between the Log2 ratios of hybridization signals and sequence identity. Subsequently, we compared the genomes of the environmental isolate NIH433 (MATa) and the clinical isolate NIH12 (MATalpha) with a tiling array of the genome of the laboratory strain JEC21 derived from these strains. In this case, CGH identified putative recombination sites and the origins of specific segments of the JEC21 genome. Similarly, CGH analysis revealed marked variability in the genomes of strains representing the VNI, VNII, and VNB molecular subtypes of the A serotype, including disomy for chromosome 13 in two strains. Additionally, CGH identified differences in chromosome content between three strains with the hybrid AD serotype and revealed that chromosome 1 from the serotype A genome is preferentially retained in all three strains.
Conclusion: The genomes of serotypes A, D, and AD strains exhibit extensive variation that spans the range from small differences (such as regions of divergence, deletion, or amplification) to the unexpected disomy for chromosome 13 in haploid strains and preferential retention of specific chromosomes in naturally occurring diploids.
Figures




Similar articles
-
Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa.PLoS Pathog. 2007 Aug 17;3(8):e114. doi: 10.1371/journal.ppat.0030114. PLoS Pathog. 2007. PMID: 17708680 Free PMC article.
-
Physical maps for genome analysis of serotype A and D strains of the fungal pathogen Cryptococcus neoformans.Genome Res. 2002 Sep;12(9):1445-53. doi: 10.1101/gr.81002. Genome Res. 2002. PMID: 12213782 Free PMC article.
-
Serotype, mating type and ploidy of Cryptococcus neoformans strains isolated from patients in Brazil.Rev Inst Med Trop Sao Paulo. 2002 Nov-Dec;44(6):299-302. doi: 10.1590/s0036-46652002000600001. Rev Inst Med Trop Sao Paulo. 2002. PMID: 12532211
-
On the origins of congenic MATalpha and MATa strains of the pathogenic yeast Cryptococcus neoformans.Fungal Genet Biol. 1999 Oct;28(1):1-5. doi: 10.1006/fgbi.1999.1155. Fungal Genet Biol. 1999. PMID: 10512666 Review.
-
Hybrids and hybridization in the Cryptococcus neoformans and Cryptococcus gattii species complexes.Infect Genet Evol. 2018 Dec;66:245-255. doi: 10.1016/j.meegid.2018.10.011. Epub 2018 Oct 17. Infect Genet Evol. 2018. PMID: 30342094 Review.
Cited by
-
Balancing stability and flexibility within the genome of the pathogen Cryptococcus neoformans.PLoS Pathog. 2013;9(12):e1003764. doi: 10.1371/journal.ppat.1003764. Epub 2013 Dec 12. PLoS Pathog. 2013. PMID: 24348244 Free PMC article. No abstract available.
-
Mitotic Recombination and Adaptive Genomic Changes in Human Pathogenic Fungi.Genes (Basel). 2019 Nov 7;10(11):901. doi: 10.3390/genes10110901. Genes (Basel). 2019. PMID: 31703352 Free PMC article. Review.
-
Clonal evolution in serially passaged Cryptococcus neoformans × deneoformans hybrids reveals a heterogenous landscape of genomic change.Genetics. 2022 Jan 4;220(1):iyab142. doi: 10.1093/genetics/iyab142. Genetics. 2022. PMID: 34849836 Free PMC article.
-
How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans.Annu Rev Microbiol. 2009;63:223-47. doi: 10.1146/annurev.micro.62.081307.162753. Annu Rev Microbiol. 2009. PMID: 19575556 Free PMC article. Review.
-
A unique chromosomal rearrangement in the Cryptococcus neoformans var. grubii type strain enhances key phenotypes associated with virulence.mBio. 2012 Feb 28;3(2):e00310-11. doi: 10.1128/mBio.00310-11. Print 2012. mBio. 2012. PMID: 22375073 Free PMC article.
References
-
- Casadevall A, Cassone A, Bistoni F, Cutler JE, Magliani W, Murphy JW, Polonelli L, Romani L. Antibody and/or cell-mediated immunity, protective mechanisms in fungal disease: an ongoing dilemma or an unnecessary dispute? Med Mycol. 1998;36(Suppl 1):95–105. - PubMed
-
- Casadevall A, Perfect JR. Cryptococcus neoformans. Washington, DC: American Society for Microbiology Press; 1998.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

