Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells
- PMID: 18294628
- PMCID: PMC2425677
- DOI: 10.1016/j.ydbio.2008.01.011
Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells
Abstract
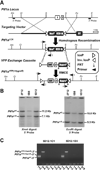
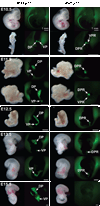
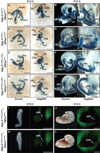
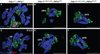
The pancreas is derived from a pool of multipotent progenitor cells (MPCs) that co-express Pdx-1 and Ptf1a. To more precisely define how the individual and combined loss of Pdx-1 and Ptf1a affects pancreatic MPC specification and differentiation we derived and studied mice bearing a novel Ptf1a(YFP) allele. While the expression of Pdx-1 and Ptf1a in pancreatic MPCs coincides between E9.5 and 12.5 the developmental phenotypes of Pdx-1 null and Pdx-1; Ptf1a double null mice are indistinguishable, and an early pancreatic bud is formed in both cases. This finding indicates that Pdx-1 is required in the foregut endoderm prior to Ptf1a for pancreatic MPC specification. We also found that Ptf1a is neither required for specification of Ngn3-positive endocrine progenitors nor differentiation of mature beta-cells. In the absence of Pdx-1 Ngn3-positive cells were not observed after E9.5. Thus, in contrast to the deletion of Ptf1a, the loss of Pdx-1 precludes the sustained Ngn3-based derivation of endocrine progenitors from pancreatic MPCs. Taken together, these studies indicate that Pdx-1 and Ptf1a have distinct but interdependent functions during pancreatic MPC specification.
Figures



References
-
- Ahlgren U, et al. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. - PubMed
-
- Apelqvist A, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. - PubMed
-
- Chiang MK, Melton DA. Single-cell transcript analysis of pancreas development. Dev Cell. 2003;4:383–393. - PubMed
-
- D'Amour KA, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

