Associative image analysis: a method for automated quantification of 3D multi-parameter images of brain tissue
- PMID: 18294697
- PMCID: PMC2700351
- DOI: 10.1016/j.jneumeth.2007.12.024
Associative image analysis: a method for automated quantification of 3D multi-parameter images of brain tissue
Abstract
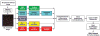
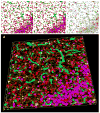
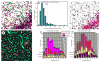
Brain structural complexity has confounded prior efforts to extract quantitative image-based measurements. We present a systematic 'divide and conquer' methodology for analyzing three-dimensional (3D) multi-parameter images of brain tissue to delineate and classify key structures, and compute quantitative associations among them. To demonstrate the method, thick ( approximately 100 microm) slices of rat brain tissue were labeled using three to five fluorescent signals, and imaged using spectral confocal microscopy and unmixing algorithms. Automated 3D segmentation and tracing algorithms were used to delineate cell nuclei, vasculature, and cell processes. From these segmentations, a set of 23 intrinsic and 8 associative image-based measurements was computed for each cell. These features were used to classify astrocytes, microglia, neurons, and endothelial cells. Associations among cells and between cells and vasculature were computed and represented as graphical networks to enable further analysis. The automated results were validated using a graphical interface that permits investigator inspection and corrective editing of each cell in 3D. Nuclear counting accuracy was >89%, and cell classification accuracy ranged from 81 to 92% depending on cell type. We present a software system named FARSIGHT implementing our methodology. Its output is a detailed XML file containing measurements that may be used for diverse quantitative hypothesis-driven and exploratory studies of the central nervous system.
Figures



References
-
- Abdul-Karim MA, Al-Kofahi K, Brown EB, Jain RK, Roysam B. Automated tracing and change analysis of angiogenic vasculature from in vivo multiphoton confocal image time series. Microvasc Res. 2003;66(2):113–25. - PubMed
-
- Al-Kofahi K, Lasek S, Szarowski DH, Dowell-Mesfin N, Shain W, Turner JN, Roysam B. Median-based robust algorithms for tracing neurons from noisy confocal microscope images. IEEE Trans Inf Technol Biomed. 2003;7:302–317. - PubMed
-
- Al-Kofahi Y, Dowell-Mesfin D, Pace C, Shain W, Turner JN, Roysam B. Improved detection od branching points in algorithms for automated neuron tracing from 3D confocal images. Cytometry A. 2007 in press. - PubMed
-
- Bear M. Neuroscience: Exploring the Brain. 3. Lippincott Williams & Wilkens; New York: 2006.
-
- Bezdek J. Pattern recognition with fuzzy objective function algorithms. Plenum; New York: 1981.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

