Large-scale screening for novel low-affinity extracellular protein interactions
- PMID: 18296487
- PMCID: PMC2279249
- DOI: 10.1101/gr.7187808
Large-scale screening for novel low-affinity extracellular protein interactions
Abstract
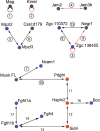
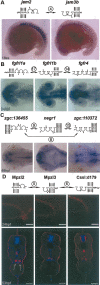
Extracellular protein-protein interactions are essential for both intercellular communication and cohesion within multicellular organisms. Approximately a fifth of human genes encode membrane-tethered or secreted proteins, but they are largely absent from recent large-scale protein interaction datasets, making current interaction networks biased and incomplete. This discrepancy is due to the unsuitability of popular high-throughput methods to detect extracellular interactions because of the biochemical intractability of membrane proteins and their interactions. For example, cell surface proteins contain insoluble hydrophobic transmembrane regions, and their extracellular interactions are often highly transient, having half-lives of less than a second. To detect transient extracellular interactions on a large scale, we developed AVEXIS (avidity-based extracellular interaction screen), a high-throughput assay that overcomes these technical issues and can detect very transient interactions (half-lives <or= 0.1 sec) with a low false-positive rate. We used it to systematically screen for receptor-ligand pairs within the zebrafish immunoglobulin superfamily and identified novel ligands for both well-known and orphan receptors. Genes encoding receptor-ligand pairs were often clustered phylogenetically and expressed in the same or adjacent tissues, immediately implying their involvement in similar biological processes. Using AVEXIS, we have determined the first systematic low-affinity extracellular protein interaction network, supported by independent biological data. This technique will now allow large-scale extracellular protein interaction mapping in a broad range of experimental contexts.
Figures



Comment in
-
A new way to explore the world of extracellular protein interactions.Genome Res. 2008 Apr;18(4):517-20. doi: 10.1101/gr.074583.107. Genome Res. 2008. PMID: 18381898
Similar articles
-
Avidity-based extracellular interaction screening (AVEXIS) for the scalable detection of low-affinity extracellular receptor-ligand interactions.J Vis Exp. 2012 Mar 5;(61):e3881. doi: 10.3791/3881. J Vis Exp. 2012. PMID: 22414956 Free PMC article.
-
A benchmarked protein microarray-based platform for the identification of novel low-affinity extracellular protein interactions.Anal Biochem. 2012 May 1;424(1):45-53. doi: 10.1016/j.ab.2012.01.034. Epub 2012 Feb 17. Anal Biochem. 2012. PMID: 22342946 Free PMC article.
-
A cell surface interaction network of neural leucine-rich repeat receptors.Genome Biol. 2009;10(9):R99. doi: 10.1186/gb-2009-10-9-r99. Epub 2009 Sep 18. Genome Biol. 2009. PMID: 19765300 Free PMC article.
-
High-throughput identification of transient extracellular protein interactions.Biochem Soc Trans. 2010 Aug;38(4):919-22. doi: 10.1042/BST0380919. Biochem Soc Trans. 2010. PMID: 20658977 Review.
-
Signal initiation in biological systems: the properties and detection of transient extracellular protein interactions.Mol Biosyst. 2009 Dec;5(12):1405-12. doi: 10.1039/B903580J. Mol Biosyst. 2009. PMID: 19593473 Free PMC article. Review.
Cited by
-
Rapid and sensitive large-scale screening of low affinity extracellular receptor protein interactions by using reaction induced inhibition of Gaussia luciferase.Sci Rep. 2020 Jun 29;10(1):10522. doi: 10.1038/s41598-020-67468-7. Sci Rep. 2020. PMID: 32601498 Free PMC article.
-
A physical wiring diagram for the human immune system.Nature. 2022 Aug;608(7922):397-404. doi: 10.1038/s41586-022-05028-x. Epub 2022 Aug 3. Nature. 2022. PMID: 35922511 Free PMC article.
-
A panel of recombinant Leishmania donovani cell surface and secreted proteins identifies LdBPK_323600.1 as a serological marker of symptomatic infection.mBio. 2024 May 8;15(5):e0085924. doi: 10.1128/mbio.00859-24. Epub 2024 Apr 19. mBio. 2024. PMID: 38639536 Free PMC article.
-
Elucidation of host-pathogen protein-protein interactions to uncover mechanisms of host cell rewiring.Curr Opin Microbiol. 2017 Oct;39:7-15. doi: 10.1016/j.mib.2017.07.005. Epub 2017 Aug 11. Curr Opin Microbiol. 2017. PMID: 28806587 Free PMC article. Review.
-
Discovery of Carbonic Anhydrase 9 as a Novel CLEC2 Ligand in a Cellular Interactome Screen.Cells. 2024 Dec 17;13(24):2083. doi: 10.3390/cells13242083. Cells. 2024. PMID: 39768175 Free PMC article.
References
-
- Aricescu A.R., Hon W.C., Siebold C., Lu W., van der Merwe P.A., Jones E.Y., Hon W.C., Siebold C., Lu W., van der Merwe P.A., Jones E.Y., Siebold C., Lu W., van der Merwe P.A., Jones E.Y., Lu W., van der Merwe P.A., Jones E.Y., van der Merwe P.A., Jones E.Y., Jones E.Y. Molecular analysis of receptor protein tyrosine phosphatase mu-mediated cell adhesion. EMBO J. 2006;25:701–712. - PMC - PubMed
-
- Barclay A.N. Membrane proteins with immunoglobulin-like domains—A master superfamily of interaction molecules. Semin. Immunol. 2003;15:215–223. - PubMed
-
- Brown M.H., Barclay A.N., Barclay A.N. Expression of immunoglobulin and scavenger receptor superfamily domains as chimeric proteins with domains 3 and 4 of CD4 for ligand analysis. Protein Eng. 1994;7:515–521. - PubMed
-
- Brown M.H., Boles K., van der Merwe P.A., Kumar V., Mathew P.A., Barclay A.N., Boles K., van der Merwe P.A., Kumar V., Mathew P.A., Barclay A.N., van der Merwe P.A., Kumar V., Mathew P.A., Barclay A.N., Kumar V., Mathew P.A., Barclay A.N., Mathew P.A., Barclay A.N., Barclay A.N. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J. Exp. Med. 1998;188:2083–2090. - PMC - PubMed
-
- Clark H.F., Gurney A.L., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Gurney A.L., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Brush J., Chen J., Chow B., Chui C., Crowley C., Chen J., Chow B., Chui C., Crowley C., Chow B., Chui C., Crowley C., Chui C., Crowley C., Crowley C., et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: A bioinformatics assessment. Genome Res. 2003;13:2265–2270. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
