Effects of SecE depletion on the inner and outer membrane proteomes of Escherichia coli
- PMID: 18296516
- PMCID: PMC2395001
- DOI: 10.1128/JB.01631-07
Effects of SecE depletion on the inner and outer membrane proteomes of Escherichia coli
Abstract
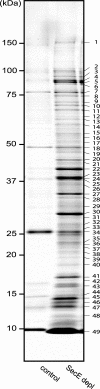
The Sec translocon is a protein-conducting channel that allows polypeptides to be transferred across or integrated into a membrane. Although protein translocation and insertion in Escherichia coli have been studied using only a small set of specific model substrates, it is generally assumed that most secretory proteins and inner membrane proteins use the Sec translocon. Therefore, we have studied the role of the Sec translocon using subproteome analysis of cells depleted of the essential translocon component SecE. The steady-state proteomes and the proteome dynamics were evaluated using one- and two-dimensional gel analysis, followed by mass spectrometry-based protein identification and extensive immunoblotting. The analysis showed that upon SecE depletion (i) secretory proteins aggregated in the cytoplasm and the cytoplasmic sigma(32) stress response was induced, (ii) the accumulation of outer membrane proteins was reduced, with the exception of OmpA, Pal, and FadL, and (iii) the accumulation of a surprisingly large number of inner membrane proteins appeared to be unaffected or increased. These proteins lacked large translocated domains and/or consisted of only one or two transmembrane segments. Our study suggests that several secretory and inner membrane proteins can use Sec translocon-independent pathways or have superior access to the remaining Sec translocons present in SecE-depleted cells.
Figures





References
-
- Akiyama, Y., A. Kihara, H. Tokuda, and K. Ito. 1996. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 27131196-31201. - PubMed
-
- Arsene, F., T. Tomoyasu, and B. Bukau. 2000. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 553-9. - PubMed
-
- Baars, L., A. J. Ytterberg, D. Drew, S. Wagner, C. Thilo, K. J. van Wijk, and J. W. de Gier. 2006. Defining the role of the Escherichia coli chaperone SecB using comparative proteomics. J. Biol. Chem. 28110024-10034. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

