Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing
- PMID: 18296532
- PMCID: PMC2293171
- DOI: 10.1128/AEM.01867-07
Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing
Abstract
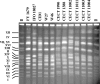
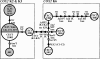
We analyzed 24 beer strains from different origins by using PCR-restriction fragment length polymorphism analysis of different gene regions, and six new Saccharomyces cerevisiae x Saccharomyces kudriavzevii hybrid strains were found. This is the first time that the presence in brewing of this new type of hybrid has been demonstrated. From the comparative molecular analysis of these natural hybrids with respect to those described in wines, it can be concluded that these originated from at least two hybridization events and that some brewing hybrids share a common origin with wine hybrids. Finally, a reduction of the S. kudriavzevii fraction of the hybrid genomes was observed, but this reduction was found to vary among hybrids regardless of the source of isolation. The fact that 25% of the strains analyzed were discovered to be S. cerevisiae x S. kudriavzevii hybrids suggests that an important fraction of brewing strains classified as S. cerevisiae may correspond to hybrids, contributing to the complexity of Saccharomyces diversity in brewing environments. The present study raises new questions about the prevalence of these new hybrids in brewing as well as their contribution to the properties of the final product.
Figures


References
-
- Bond, U., and A. Blomberg. 2006. Principles and applications of genomics and proteomics in the analysis of industrial yeast strains, p. 175-214. In A. Querol and G. H. Fleet (ed.), Yeasts in food and beverages. Springer-Verlag, Berlin, Germany.
-
- Bond, U., C. Neal, D. Donnelly, and T. C. James. 2004. Aneuploidy and copy number breakpoints in the genome of lager yeasts mapped by microarray hybridisation. Curr. Genet. 45:360-370. - PubMed
-
- Bradbury, J., K. Richards, H. Niederer, S. Lee, P. Rod Dunbar, and R. Gardner. 2006. A homozygous diploid subset of commercial wine yeast strains. Antonie van Leeuwenhoek 89:27-37. - PubMed
-
- Casaregola, S., H. V. Nguyen, G. Lapathitis, A. Kotyk, and C. Gaillardin. 2001. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int. J. Syst. Evol. Microbiol. 51:1607-1618. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

