Large-scale chemical dissection of mitochondrial function
- PMID: 18297058
- PMCID: PMC2715872
- DOI: 10.1038/nbt1387
Large-scale chemical dissection of mitochondrial function
Erratum in
- Nat Biotechnol. 2008 Jul;26(7):831
Abstract
Mitochondrial oxidative phosphorylation (OXPHOS) is under the control of both mitochondrial (mtDNA) and nuclear genomes and is central to energy homeostasis. To investigate how its function and regulation are integrated within cells, we systematically combined four cell-based assays of OXPHOS physiology with multiplexed measurements of nuclear and mtDNA gene expression across 2,490 small-molecule perturbations in cultured muscle. Mining the resulting compendium revealed, first, that protein synthesis inhibitors can decouple coordination of nuclear and mtDNA transcription; second, that a subset of HMG-CoA reductase inhibitors, combined with propranolol, can cause mitochondrial toxicity, yielding potential clues about the etiology of statin myopathy; and, third, that structurally diverse microtubule inhibitors stimulate OXPHOS transcription while suppressing reactive oxygen species, via a transcriptional mechanism involving PGC-1alpha and ERRalpha, and thus may be useful in treating age-associated degenerative disorders. Our screening compendium can be used as a discovery tool both for understanding mitochondrial biology and toxicity and for identifying novel therapeutics.
Figures


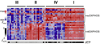


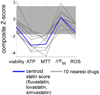

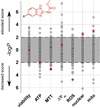





Comment in
-
A toolkit for the cell's powerhouse.Nat Biotechnol. 2008 Mar;26(3):294-6. doi: 10.1038/nbt0308-294. Nat Biotechnol. 2008. PMID: 18327238 No abstract available.
References
-
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. III. The steady state. J. Biol. Chem. 1955;217:409–427. - PubMed
-
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. - PubMed
-
- Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

