Quantification of Cre-mediated recombination by a novel strategy reveals a stable extra-chromosomal deletion-circle in mice
- PMID: 18298805
- PMCID: PMC2277394
- DOI: 10.1186/1472-6750-8-18
Quantification of Cre-mediated recombination by a novel strategy reveals a stable extra-chromosomal deletion-circle in mice
Abstract
Background: Inducible conditional knockout animals are widely used to get insight in the function of genes and the pathogenesis of human diseases. These models frequently rely on Cre-mediated recombination of sequences flanked by Lox-P sites. To understand the consequences of gene disruption, it is essential to know the efficiency of the recombination process.
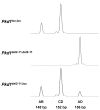
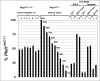
Results: Here, we describe a modification of the multiplex ligation-dependent probe amplification (MLPA), called extension-MLPA (eMLPA), which enables quantification of relatively small differences in DNA that are a consequence of Cre-mediated recombination. eMLPA, here applied on an inducible Pkd1 conditional deletion mouse model, simultaneously measures both the reduction of the floxed allele and the increase of the deletion allele in a single reaction thereby minimizing any type of experimental variation. Interestingly, with this method we were also able to observe the presence of the excised DNA fragment. This extra-chromosomal deletion-circle was detectable up to 5 months after activation of Cre.
Conclusion: eMLPA is a novel strategy which easily can be applied to measure the Cre-mediated recombination efficiency in each experimental case with high accuracy. In addition the fate of the deletion-circle can be followed simultaneously.
Figures




References
-
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

