Gene conversion in the rice genome
- PMID: 18298833
- PMCID: PMC2277409
- DOI: 10.1186/1471-2164-9-93
Gene conversion in the rice genome
Abstract
Background: Gene conversion causes a non-reciprocal transfer of genetic information between similar sequences. Gene conversion can both homogenize genes and recruit point mutations thereby shaping the evolution of multigene families. In the rice genome, the large number of duplicated genes increases opportunities for gene conversion.
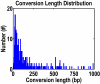
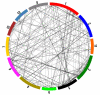
Results: To characterize gene conversion in rice, we have defined 626 multigene families in which 377 gene conversions were detected using the GENECONV program. Over 60% of the conversions we detected were between chromosomes. We found that the inter-chromosomal conversions distributed between chromosome 1 and 5, 2 and 6, and 3 and 5 are more frequent than genome average (Z-test, P < 0.05). The frequencies of gene conversion on the same chromosome decreased with the physical distance between gene conversion partners. Ka/Ks analysis indicates that gene conversion is not tightly linked to natural selection in the rice genome. To assess the contribution of segmental duplication on gene conversion statistics, we determined locations of conversion partners with respect to inter-chromosomal segment duplication. The number of conversions associated with segmentation is less than ten percent. Pseudogenes in the rice genome with low similarity to Arabidopsis genes showed greater likelihood for gene conversion than those with high similarity to Arabidopsis genes. Functional annotations suggest that at least 14 multigene families related to disease or bacteria resistance were involved in conversion events.
Conclusion: The evolution of gene families in the rice genome may have been accelerated by conversion with pseudogenes. Our analysis suggests a possible role for gene conversion in the evolution of pathogen-response genes.
Figures


Similar articles
-
Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining.BMC Genomics. 2008 Feb 21;9:86. doi: 10.1186/1471-2164-9-86. BMC Genomics. 2008. PMID: 18291034 Free PMC article.
-
Conversion between 100-million-year-old duplicated genes contributes to rice subspecies divergence.BMC Genomics. 2021 Jun 19;22(1):460. doi: 10.1186/s12864-021-07776-y. BMC Genomics. 2021. PMID: 34147070 Free PMC article.
-
Extensive concerted evolution of rice paralogs and the road to regaining independence.Genetics. 2007 Nov;177(3):1753-63. doi: 10.1534/genetics.107.073197. Genetics. 2007. PMID: 18039882 Free PMC article.
-
Ectopic gene conversions in the human genome.Genomics. 2009 Jan;93(1):27-32. doi: 10.1016/j.ygeno.2008.09.007. Epub 2008 Oct 19. Genomics. 2009. PMID: 18848875 Review.
-
Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species.Plant Physiol. 2004 Jun;135(2):622-9. doi: 10.1104/pp.104.040170. Plant Physiol. 2004. PMID: 15208410 Free PMC article. Review.
Cited by
-
Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster.Genome Res. 2017 May;27(5):709-721. doi: 10.1101/gr.213512.116. Epub 2017 Apr 3. Genome Res. 2017. PMID: 28373483 Free PMC article.
-
Gene Conversion-Like Events in the Diversification of Human Rearranged IGHV3-23*01 Gene Sequences.Front Immunol. 2012 Jun 15;3:158. doi: 10.3389/fimmu.2012.00158. eCollection 2012. Front Immunol. 2012. PMID: 22715339 Free PMC article.
-
Nonallelic gene conversion in the genus Drosophila.Genetics. 2010 May;185(1):95-103. doi: 10.1534/genetics.110.115444. Epub 2010 Mar 9. Genetics. 2010. PMID: 20215470 Free PMC article.
-
Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays.BMC Plant Biol. 2010 Oct 21;10:226. doi: 10.1186/1471-2229-10-226. BMC Plant Biol. 2010. PMID: 20964856 Free PMC article.
-
Insights into the evolution of cotton diploids and polyploids from whole-genome re-sequencing.G3 (Bethesda). 2013 Oct 3;3(10):1809-18. doi: 10.1534/g3.113.007229. G3 (Bethesda). 2013. PMID: 23979935 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

