Specificity of DNA microarray hybridization: characterization, effectors and approaches for data correction
- PMID: 18299281
- PMCID: PMC2367720
- DOI: 10.1093/nar/gkn087
Specificity of DNA microarray hybridization: characterization, effectors and approaches for data correction
Abstract
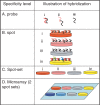
Microarray-hybridization specificity is one of the main effectors of microarray result quality. In the present review, we suggest a definition for specificity that spans four hybridization levels, from the single probe to the microarray platform. For increased hybridization specificity, it is important to quantify the extent of the specificity at each of these levels, and correct the data accordingly. We outline possible effects of low hybridization specificity on the obtained results and list possible effectors of hybridization specificity. In addition, we discuss several studies in which theoretical approaches, empirical means or data filtration were used to identify specificity effectors, and increase the specificity of the hybridization results. However, these various approaches may not yet provide an ultimate solution; rather, further tool development is needed to enhance microarray-hybridization specificity.
Figures

References
-
- Stoughton RB. Applications of DNA microarrays in biology. Annu. Rev. Biochem. 2005;74:53–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

