Requirements for gene silencing mediated by U1 snRNA binding to a target sequence
- PMID: 18299285
- PMCID: PMC2367729
- DOI: 10.1093/nar/gkn068
Requirements for gene silencing mediated by U1 snRNA binding to a target sequence
Abstract
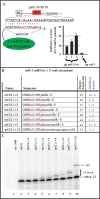
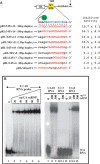
U1 interference (U1i) is a novel method to block gene expression. U1i requires expression of a 5'-end-mutated U1 snRNA designed to base pair to the 3'-terminal exon of the target gene's pre-mRNA that leads to inhibition of polyadenylation. Here, we show U1i is robust (> or =95%) and a 10-nt target length is sufficient for good silencing. Surprisingly, longer U1 snRNAs, which could increase annealing to the target, fail to improve silencing. Extensive mutagenesis of the 10-bp U1 snRNA:target duplex shows that any single mismatch different from GU at positions 3-8, destroys silencing. However, mismatches within the other positions give partial silencing, suggesting that off-target inhibition could occur. The specificity of U1i may be enhanced, however, by the fact that silencing is impaired by RNA secondary structure or by splicing factors binding nearby, the latter mediated by Arginine-Serine (RS) domains. U1i inhibition can be reconstituted in vivo by tethering of RS domains of U1-70K and U2AF65. These results help to: (i) define good target sites for U1i; (ii) identify and understand natural cellular examples of U1i; (iii) clarify the contribution of hydrogen bonding to U1i and to U1 snRNP binding to 5' splice sites and (iv) understand the mechanism of U1i.
Figures




References
-
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

