Human cytidine deaminase APOBEC3H restricts HIV-1 replication
- PMID: 18299330
- PMCID: PMC2430661
- DOI: 10.1074/jbc.M707586200
Human cytidine deaminase APOBEC3H restricts HIV-1 replication
Abstract
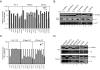
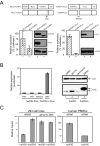
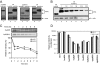
The human genome encodes seven APOBEC3 (A3) cytidine deaminases with potential antiretroviral activity: A3A, A3B, A3C, A3DE, A3F, A3G, and A3H. A3G was the first identified to block replication of human immunodeficiency virus type 1 (HIV-1) and many other retroviruses. A3F, A3B, and A3DE were shown later to have similar activities. HIV-1 produces a protein called Vif that is able to neutralize the antiretroviral activities of A3DE, A3F, and A3G, but not A3B. Only the antiretroviral activity of A3H remains to be defined due to its poor expression in cell culture. Here, we studied the mechanism impairing A3H expression. When primate A3H sequences were compared, a premature termination codon was identified on the fifth exon of the human and chimpanzee A3H genes, which significantly decreased their protein expression. It causes a 29-residue deletion from the C terminus, and this truncation did not reduce human A3H protein stability. However, the mRNA levels of the truncated gene were significantly decreased. Human A3H protein expression could be restored to a normal level either by repairing this truncation or through expression from a vector containing an intron from human cytomegalovirus. Once expression was optimized, human A3H could reduce HIV-1 infectivity up to 150-fold. Importantly, HIV-1 Vif failed to neutralize A3H activity. Nevertheless, extensive sequence analysis could not detect any significant levels of G-to-A mutation in the HIV-1 genome by human A3H. Thus, A3H inhibits HIV-1 replication potently by a cytidine deamination-independent mechanism, and optimizing A3H expression in vivo should represent a novel therapeutic strategy for HIV-1 treatment.
Figures


References
-
- Holmes, R. K., Malim, M. H., and Bishop, K. N. (2007) Trends Biochem. Sci. 32 118-128 - PubMed
-
- Bishop, K. N., Holmes, R. K., Sheehy, A. M., Davidson, N. O., Cho, S. J., and Malim, M. H. (2004) Curr. Biol. 14 1392-1396 - PubMed
-
- Doehle, B. P., Schafer, A., and Cullen, B. R. (2005) Virology 339 281-288 - PubMed
-
- Liddament, M. T., Brown, W. L., Schumacher, A. J., and Harris, R. S. (2004) Curr. Biol. 14 1385-1391 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

