Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system
- PMID: 18299388
- PMCID: PMC2423155
- DOI: 10.1128/MCB.01469-07
Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system
Abstract
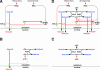
The NF-kappaB signaling pathway regulates the activity of multiple dimeric transcription factors that are generated from five distinct monomers. The availabilities of specific dimers are regulated during cell differentiation and organ development and determine the cell's responsiveness to inflammatory or developmental signals. An altered dimer distribution is a hallmark of many chronic diseases. Here, we reveal that the cellular processes that generate different NF-kappaB dimers are highly connected through multiple cross-regulatory mechanisms. First, we find that steady-state expression of RelB is regulated by the canonical pathway and constitutive RelA activity. Indeed, synthesis control of RelB is the major determinant of noncanonical NF-kappaB dimer activation. Second, processing, not synthesis, of p100 and p105 is mechanistically linked via competitive dimerization with a limited pool of RelA and RelB. This homeostatic cross-regulatory mechanism determines the availability of the p50- and p52-containing dimers and also of the noncanonical IkappaB p100. Our results inform a wiring diagram to delineate NF-kappaB dimer formation that emphasizes that inflammatory and developmental signaling cannot be considered separately but are highly interconnected.
Figures
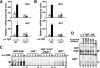






References
-
- Bonizzi, G., and M. Karin. 2004. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25280-288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
