Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers
- PMID: 18299561
- PMCID: PMC2265152
- DOI: 10.1073/pnas.0712384105
Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers
Abstract
Although the majority of colorectal cancers exhibit chromosome instability (CIN), only a few genes that might cause this phenotype have been identified and no general mechanism underlying their function has emerged. To systematically identify somatic mutations in potential CIN genes in colorectal cancers, we determined the sequence of 102 human homologues of 96 yeast CIN genes known to function in various aspects of chromosome transmission fidelity. We identified 11 somatic mutations distributed among five genes in a panel that included 132 colorectal cancers. Remarkably, all but one of these 11 mutations were in the homologs of yeast genes that regulate sister chromatid cohesion. We then demonstrated that down-regulation of such homologs resulted in chromosomal instability and chromatid cohesion defects in human cells. Finally, we showed that down-regulation or genetic disruption of the two major candidate CIN genes identified in previous studies (MRE11A and CDC4) also resulted in abnormal sister chromatid cohesion in human cells. These results suggest that defective sister chromatid cohesion as a result of somatic mutations may represent a major cause of chromosome instability in human cancers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

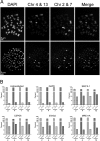
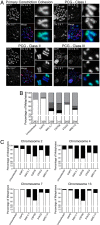

References
-
- Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. - PubMed
-
- Duesberg P, et al. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 1999;19:4887–4906. - PubMed
-
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. - PubMed
-
- Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. - PubMed
-
- Yamamoto H, Imai K, Perucho M. Gastrointestinal cancer of the microsatellite mutator phenotype pathway. J Gastroenterol. 2002;37:153–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

