Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase
- PMID: 18301062
- PMCID: PMC2921824
- DOI: 10.1097/QAD.0b013e3282f29478
Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase
Abstract
Background: Failure of antiretroviral therapy may result from the selection of pre-existing, drug-resistant HIV-1 variants, but the frequency and type of such variants have not been defined.
Objective: We used single genome sequencing (SGS) to characterize the frequency of polymorphism at drug resistance sites in protease (PR) and reverse transcriptase (RT) in plasma samples from antiretroviral naive individuals.
Methods: A total of 2229 pro-pol sequences in 79 plasma samples from 30 patients were analyzed by SGS. A mean of 28 single genome sequences was obtained from each sample. The frequency of mutations at all PR and RT sites was compared to those associated with drug resistance.
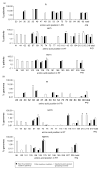
Results: We detected polymorphism at one or more drug resistance sites in 27 of 30 (90%) patients. Polymorphism at positions 179 and 215 of RT was most common, both occurring in 23% of patients. Most (68%) of other drug resistance sites were polymorphic with an average of 3.2% of genomes per sample containing at least one variant from wild type. Seven drug resistance sites were polymorphic in more than 1% of genomes: PR position 33; RT positions 69, 98, 118, 179, 210, and 215. Although frequencies of synonymous polymorphism were similar at resistance and nonresistance sites, nonsynonymous polymorphism were significantly less common at drug resistance sites, implying stronger purifying selection at these positions.
Conclusions: HIV-1 variants that are polymorphic at drug resistance sites pre-exist frequently as minor species in antiretroviral naive individuals. Standard genotype techniques have grossly underestimated their frequency.
Figures

References
-
- Palmer S, Boltz V, Maldarelli F, Kearney M, Halvas EK, Rock D, et al. Selection and persistence of nonnucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping nonnucleoside therapy. Aids. 2006;20:701–710. - PubMed
-
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

