Mapping of immunogenic and protein-interacting regions at the surface of the seven-bladed beta-propeller domain of the HIV-1 cellular interactor EED
- PMID: 18302803
- PMCID: PMC2292171
- DOI: 10.1186/1743-422X-5-32
Mapping of immunogenic and protein-interacting regions at the surface of the seven-bladed beta-propeller domain of the HIV-1 cellular interactor EED
Abstract
Background: The human EED protein, a member of the superfamily of Polycomb group proteins, is involved in multiple cellular protein complexes. Its C-terminal domain, which is common to the four EED isoforms, contains seven repeats of a canonical WD-40 motif. EED is an interactor of three HIV-1 proteins, matrix (MA), integrase (IN) and Nef. An antiviral activity has been found to be associated with isoforms EED3 and EED4 at the late stage of HIV-1 replication, due to a negative effect on virus assembly and genomic RNA packaging. The aim of the present study was to determine the regions of the EED C-terminal core domain which were accessible and available to protein interactions, using three-dimensional (3D) protein homology modelling with a WD-40 protein of known structure, and epitope mapping of anti-EED antibodies.
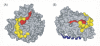
Results: Our data suggested that the C-terminal domain of EED was folded as a seven-bladed beta-propeller protein. During the completion of our work, crystallographic data of EED became available from co-crystals of the EED C-terminal core with the N-terminal domain of its cellular partner EZH2. Our 3D-model was in good congruence with the refined structural model determined from crystallographic data, except for a unique alpha-helix in the fourth beta-blade. More importantly, the position of flexible loops and accessible beta-strands on the beta-propeller was consistent with our mapping of immunogenic epitopes and sites of interaction with HIV-1 MA and IN. Certain immunoreactive regions were found to overlap with the EZH2, MA and IN binding sites, confirming their accessibility and reactivity at the surface of EED. Crystal structure of EED showed that the two discrete regions of interaction with MA and IN did not overlap with each other, nor with the EZH2 binding pocket, but were contiguous, and formed a continuous binding groove running along the lateral face of the beta-propeller.
Conclusion: Identification of antibody-, MA-, IN- and EZH2-binding sites at the surface of the EED isoform 3 provided a global picture of the immunogenic and protein-protein interacting regions in the EED C-terminal domain, organized as a seven-bladed beta-propeller protein. Mapping of the HIV-1 MA and IN binding sites on the 3D-model of EED core predicted that EED-bound MA and IN ligands would be in close vicinity at the surface of the beta-propeller, and that the occurrence of a ternary complex MA-EED-IN would be possible.
Figures


References
-
- Sewalt RGAB, van der Vlag J, Gunster MJ, Hamer KM, den Blaauwen JL, Satijn DPE, Hendrix T, van Driel R, Otte AP. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

