Flexible ligand docking to multiple receptor conformations: a practical alternative
- PMID: 18302984
- PMCID: PMC2396190
- DOI: 10.1016/j.sbi.2008.01.004
Flexible ligand docking to multiple receptor conformations: a practical alternative
Abstract
State of the art docking algorithms predict an incorrect binding pose for about 50-70% of all ligands when only a single fixed receptor conformation is considered. In many more cases, lack of receptor flexibility results in meaningless ligand binding scores, even when the correct pose is obtained. Incorporating conformational rearrangements of the receptor binding pocket into predictions of both ligand binding pose and binding score is crucial for improving structure-based drug design and virtual ligand screening methodologies. However, direct modeling of protein binding site flexibility remains challenging because of the large conformational space that must be sampled, and difficulties remain in constructing a suitably accurate energy function. Here we show that using multiple fixed receptor conformations, either experimentally determined by crystallography or NMR, or computationally generated, is a practical shortcut that may improve docking calculations. In several cases, such an approach has led to experimentally validated predictions.
Figures
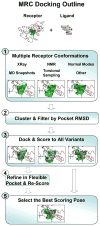
References
-
- Teodoro ML, Kavraki LE. Conformational flexibility models for the receptor in structure based drug design. Current Pharmaceutical Design. 2003;9:1635–1648. - PubMed
-
*This excellent review classifies approaches to the induced fit docking into five categories.
-
- Totrov M, Abagyan R. Detailed Ab-Initio Prediction of Lysozyme-Antibody Complex with 1.6-Angstrom Accuracy. Nat Struct Biol. 1994;1:259–263. - PubMed
-
- Yang AYC, Kallblad P, Mancera RL. Molecular modelling prediction of ligand binding site flexibility. Journal of Computer-Aided Molecular Design. 2004;18:235–250. - PubMed
-
- Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

