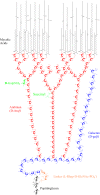
The identification and location of succinyl residues and the characterization of the interior arabinan region allow for a model of the complete primary structure of Mycobacterium tuberculosis mycolyl arabinogalactan
- PMID: 18303028
- PMCID: PMC2442352
- DOI: 10.1074/jbc.M800222200
The identification and location of succinyl residues and the characterization of the interior arabinan region allow for a model of the complete primary structure of Mycobacterium tuberculosis mycolyl arabinogalactan
Abstract
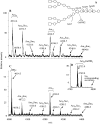
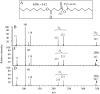
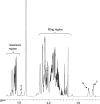
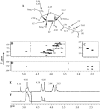
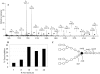
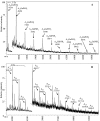
The complex cell wall of Mycobacterium tuberculosis is the hallmark of acid fast bacteria and is responsible for much of its physiological characteristics. Hence, much effort has been made to determine its primary structure. Such studies have been hampered by its extreme complexity. Also, its insolubility leads to difficulties determining the presence or absence of base labile groups. We have used an endogenous arabinase to solubilize the arabinan region of the cell wall and have shown using mass spectrometry and NMR that succinyl esters are present on O2 of the inner-branched 1,3,5-alpha-d-arabinofuranosyl residues. In addition, an inner arabinan region of 14 linear alpha-1,5 arabinofuranosyl residues has been identified. These and earlier results now allow the presentation of a model of the entire primary structure of the mycobacterial mycolyl arabinogalactan highlighted by three arabinan chains of 31 residues each.
Figures
References
-
- Kanetsuna, F. (1968) Biochim. Biophys. Acta 158 130–143 - PubMed
-
- Wietzerbin, J., Das, B. C., Petit, J. F., Lederer, E., Leyh Bouille, M., and Ghuysen, J. M. (1974) Biochemistry 13 3471–3476 - PubMed
-
- Dmitriev, B. A., Ehlers, S., Rietschel, E. T., and Brennan, P. J. (2000) Int. J. Med. Microbiol. 290 251–258 - PubMed
-
- Minnikin, D. E. (1982) in The Biology of the Mycobacteria (Ratledge, C., and Stanford, J. L., eds) Vol. I, pp. 95–184, Academic Press, London
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

