The evolution of combinatorial gene regulation in fungi
- PMID: 18303948
- PMCID: PMC2253631
- DOI: 10.1371/journal.pbio.0060038
The evolution of combinatorial gene regulation in fungi
Abstract
It is widely suspected that gene regulatory networks are highly plastic. The rapid turnover of transcription factor binding sites has been predicted on theoretical grounds and has been experimentally demonstrated in closely related species. We combined experimental approaches with comparative genomics to focus on the role of combinatorial control in the evolution of a large transcriptional circuit in the fungal lineage. Our study centers on Mcm1, a transcriptional regulator that, in combination with five cofactors, binds roughly 4% of the genes in Saccharomyces cerevisiae and regulates processes ranging from the cell-cycle to mating. In Kluyveromyces lactis and Candida albicans, two other hemiascomycetes, we find that the Mcm1 combinatorial circuits are substantially different. This massive rewiring of the Mcm1 circuitry has involved both substantial gain and loss of targets in ancient combinatorial circuits as well as the formation of new combinatorial interactions. We have dissected the gains and losses on the global level into subsets of functionally and temporally related changes. One particularly dramatic change is the acquisition of Mcm1 binding sites in close proximity to Rap1 binding sites at 70 ribosomal protein genes in the K. lactis lineage. Another intriguing and very recent gain occurs in the C. albicans lineage, where Mcm1 is found to bind in combination with the regulator Wor1 at many genes that function in processes associated with adaptation to the human host, including the white-opaque epigenetic switch. The large turnover of Mcm1 binding sites and the evolution of new Mcm1-cofactor interactions illuminate in sharp detail the rapid evolution of combinatorial transcription networks.
Conflict of interest statement
Figures



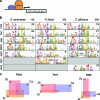

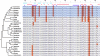

Similar articles
-
Intrinsic cooperativity potentiates parallel cis-regulatory evolution.Elife. 2018 Sep 10;7:e37563. doi: 10.7554/eLife.37563. Elife. 2018. PMID: 30198843 Free PMC article.
-
Interactions of the Mcm1 MADS box protein with cofactors that regulate mating in yeast.Mol Cell Biol. 2002 Jul;22(13):4607-21. doi: 10.1128/MCB.22.13.4607-4621.2002. Mol Cell Biol. 2002. PMID: 12052870 Free PMC article.
-
Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae.Mol Cell Biol. 1995 Nov;15(11):5917-28. doi: 10.1128/MCB.15.11.5917. Mol Cell Biol. 1995. PMID: 7565744 Free PMC article.
-
The SRF and MCM1 transcription factors.Curr Opin Genet Dev. 1992 Apr;2(2):221-6. doi: 10.1016/s0959-437x(05)80277-1. Curr Opin Genet Dev. 1992. PMID: 1638115 Review.
-
Evolution of transcription networks--lessons from yeasts.Curr Biol. 2010 Sep 14;20(17):R746-53. doi: 10.1016/j.cub.2010.06.056. Curr Biol. 2010. PMID: 20833319 Free PMC article. Review.
Cited by
-
Mutational robustness of gene regulatory networks.PLoS One. 2012;7(1):e30591. doi: 10.1371/journal.pone.0030591. Epub 2012 Jan 25. PLoS One. 2012. PMID: 22295094 Free PMC article.
-
A phenotypic profile of the Candida albicans regulatory network.PLoS Genet. 2009 Dec;5(12):e1000783. doi: 10.1371/journal.pgen.1000783. Epub 2009 Dec 24. PLoS Genet. 2009. PMID: 20041210 Free PMC article.
-
Intrinsic cooperativity potentiates parallel cis-regulatory evolution.Elife. 2018 Sep 10;7:e37563. doi: 10.7554/eLife.37563. Elife. 2018. PMID: 30198843 Free PMC article.
-
Use with caution: developmental systems divergence and potential pitfalls of animal models.Yale J Biol Med. 2009 Jun;82(2):53-66. Yale J Biol Med. 2009. PMID: 19562005 Free PMC article. Review.
-
Towards an evolutionary model of transcription networks.PLoS Comput Biol. 2011 Jun;7(6):e1002064. doi: 10.1371/journal.pcbi.1002064. Epub 2011 Jun 9. PLoS Comput Biol. 2011. PMID: 21695281 Free PMC article.
References
-
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–138. - PubMed
-
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
-
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. - PubMed
-
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: molecular genetics and the evolution of animal design. Malden (Massachussets): Blackwell Pub; 2005. 258 ix,
-
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

