Recent advances in implicit solvent-based methods for biomolecular simulations
- PMID: 18304802
- PMCID: PMC2386893
- DOI: 10.1016/j.sbi.2008.01.003
Recent advances in implicit solvent-based methods for biomolecular simulations
Abstract
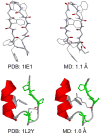
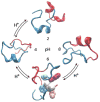
Implicit solvent-based methods play an increasingly important role in molecular modeling of biomolecular structure and dynamics. Recent methodological developments have mainly focused on the extension of the generalized Born (GB) formalism for variable dielectric environments and accurate treatment of nonpolar solvation. Extensive efforts in parameterization of GB models and implicit solvent force fields have enabled ab initio simulation of protein folding to native or near-native structures. Another exciting area that has benefited from the advances in implicit solvent models is the development of constant pH molecular dynamics methods, which have recently been applied to the calculations of protein pK(a) values and the studies of pH-dependent peptide and protein folding.
Figures
Similar articles
-
The effects of implicit modeling of nonpolar solvation on protein folding simulations.Phys Chem Chem Phys. 2018 Jul 11;20(27):18410-18419. doi: 10.1039/c8cp03156h. Phys Chem Chem Phys. 2018. PMID: 29946610
-
Generalized Born Implicit Solvent Models for Biomolecules.Annu Rev Biophys. 2019 May 6;48:275-296. doi: 10.1146/annurev-biophys-052118-115325. Epub 2019 Mar 11. Annu Rev Biophys. 2019. PMID: 30857399 Free PMC article. Review.
-
Recent advances in the development and application of implicit solvent models in biomolecule simulations.Curr Opin Struct Biol. 2004 Apr;14(2):217-24. doi: 10.1016/j.sbi.2004.03.009. Curr Opin Struct Biol. 2004. PMID: 15093837 Review.
-
A generalized Kirkwood implicit solvent for the polarizable AMOEBA protein model.J Chem Phys. 2023 Aug 7;159(5):054102. doi: 10.1063/5.0158914. J Chem Phys. 2023. PMID: 37526158 Free PMC article.
-
Secondary structure bias in generalized Born solvent models: comparison of conformational ensembles and free energy of solvent polarization from explicit and implicit solvation.J Phys Chem B. 2007 Feb 22;111(7):1846-57. doi: 10.1021/jp066831u. Epub 2007 Jan 27. J Phys Chem B. 2007. PMID: 17256983 Free PMC article.
Cited by
-
Level-Set Variational Implicit-Solvent Modeling of Biomolecules with the Coulomb-Field Approximation.J Chem Theory Comput. 2012 Feb 14;8(2):386-397. doi: 10.1021/ct200647j. Epub 2011 Dec 19. J Chem Theory Comput. 2012. PMID: 22346739 Free PMC article.
-
Guardians of the Cell: State-of-the-Art of Membrane Proteins from a Computational Point-of-View.Methods Mol Biol. 2021;2315:3-28. doi: 10.1007/978-1-0716-1468-6_1. Methods Mol Biol. 2021. PMID: 34302667
-
Connecting free energy surfaces in implicit and explicit solvent: an efficient method to compute conformational and solvation free energies.J Chem Theory Comput. 2015 Jun 9;11(6):2868-78. doi: 10.1021/acs.jctc.5b00264. J Chem Theory Comput. 2015. PMID: 26236174 Free PMC article.
-
Analysis of fast boundary-integral approximations for modeling electrostatic contributions of molecular binding.Mol Based Math Biol. 2013 Jun;1:124-150. doi: 10.2478/mlbmb-2013-0007. Mol Based Math Biol. 2013. PMID: 24466561 Free PMC article.
-
Ionic solvation studied by image-charge reaction field method.J Chem Phys. 2011 Jan 28;134(4):044105. doi: 10.1063/1.3530094. J Chem Phys. 2011. PMID: 21280685 Free PMC article.
References
-
- Feig M, Brooks CL., III Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14:217–224. - PubMed
-
- Baker NA. Improving implicit solvent simulations: a poisson-centric view. Curr Opin Struct Biol. 2005;15:137–143. - PubMed
-
- Koehl P. Electrostatics calculations: latest methodological advances. Curr Opin Struct Biol. 2006;16:142–151. - PubMed
-
- Mongan John, Case David A. Biomolecular simulations at constant pH. Curr Opin Struct Biol. 2005;15:157–163. - PubMed
-
- Ferrara P, Apostolakis J, Caflisch A. Evaluation of a fast implicit solvent model for molecular dynamics simulations. Proteins. 2002;46:24–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

