Recent advances in implicit solvent-based methods for biomolecular simulations
- PMID: 18304802
- PMCID: PMC2386893
- DOI: 10.1016/j.sbi.2008.01.003
Recent advances in implicit solvent-based methods for biomolecular simulations
Abstract
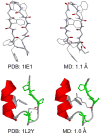
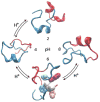
Implicit solvent-based methods play an increasingly important role in molecular modeling of biomolecular structure and dynamics. Recent methodological developments have mainly focused on the extension of the generalized Born (GB) formalism for variable dielectric environments and accurate treatment of nonpolar solvation. Extensive efforts in parameterization of GB models and implicit solvent force fields have enabled ab initio simulation of protein folding to native or near-native structures. Another exciting area that has benefited from the advances in implicit solvent models is the development of constant pH molecular dynamics methods, which have recently been applied to the calculations of protein pK(a) values and the studies of pH-dependent peptide and protein folding.
Figures
References
-
- Feig M, Brooks CL., III Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14:217–224. - PubMed
-
- Baker NA. Improving implicit solvent simulations: a poisson-centric view. Curr Opin Struct Biol. 2005;15:137–143. - PubMed
-
- Koehl P. Electrostatics calculations: latest methodological advances. Curr Opin Struct Biol. 2006;16:142–151. - PubMed
-
- Mongan John, Case David A. Biomolecular simulations at constant pH. Curr Opin Struct Biol. 2005;15:157–163. - PubMed
-
- Ferrara P, Apostolakis J, Caflisch A. Evaluation of a fast implicit solvent model for molecular dynamics simulations. Proteins. 2002;46:24–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

