Competition and coexistence of sulfate-reducing bacteria, acetogens and methanogens in a lab-scale anaerobic bioreactor as affected by changing substrate to sulfate ratio
- PMID: 18305937
- PMCID: PMC2271084
- DOI: 10.1007/s00253-008-1391-8
Competition and coexistence of sulfate-reducing bacteria, acetogens and methanogens in a lab-scale anaerobic bioreactor as affected by changing substrate to sulfate ratio
Abstract
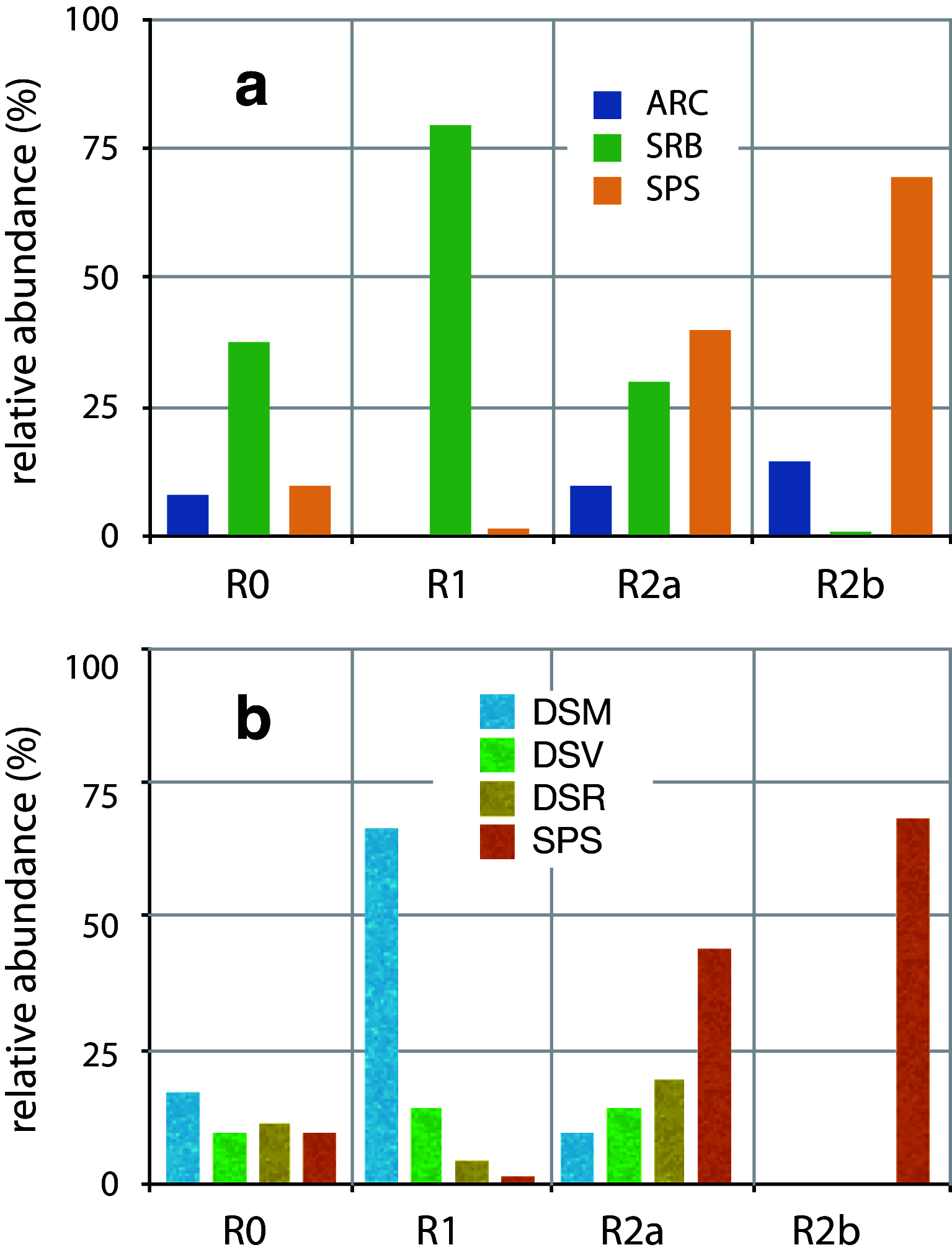
The microbial population structure and function of natural anaerobic communities maintained in lab-scale continuously stirred tank reactors at different lactate to sulfate ratios and in the absence of sulfate were analyzed using an integrated approach of molecular techniques and chemical analysis. The population structure, determined by denaturing gradient gel electrophoresis and by the use of oligonucleotide probes, was linked to the functional changes in the reactors. At the influent lactate to sulfate molar ratio of 0.35 mol mol(-1), i.e., electron donor limitation, lactate oxidation was mainly carried out by incompletely oxidizing sulfate-reducing bacteria, which formed 80-85% of the total bacterial population. Desulfomicrobium- and Desulfovibrio-like species were the most abundant sulfate-reducing bacteria. Acetogens and methanogenic Archaea were mostly outcompeted, although less than 2% of an acetogenic population could still be observed at this limiting concentration of lactate. In the near absence of sulfate (i.e., at very high lactate/sulfate ratio), acetogens and methanogenic Archaea were the dominant microbial communities. Acetogenic bacteria represented by Dendrosporobacter quercicolus-like species formed more than 70% of the population, while methanogenic bacteria related to uncultured Archaea comprising about 10-15% of the microbial community. At an influent lactate to sulfate molar ratio of 2 mol mol(-1), i.e., under sulfate-limiting conditions, a different metabolic route was followed by the mixed anaerobic community. Apparently, lactate was fermented to acetate and propionate, while the majority of sulfidogenesis and methanogenesis were dependent on these fermentation products. This was consistent with the presence of significant levels (40-45% of total bacteria) of D. quercicolus-like heteroacetogens and a corresponding increase of propionate-oxidizing Desulfobulbus-like sulfate-reducing bacteria (20% of the total bacteria). Methanogenic Archaea accounted for 10% of the total microbial community.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

