Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules
- PMID: 18308592
- PMCID: PMC2287197
- DOI: 10.1016/j.immuni.2008.01.008
Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules
Abstract
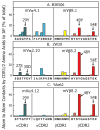
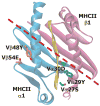
To test whether highly crossreactive alphabeta T cell receptors (TCRs) produced during limited negative selection best illustrate evolutionarily conserved interactions between TCR and major histocompatibility complex (MHC) molecules, we solved the structures of three TCRs bound to the same MHC II peptide (IAb-3K). The TCRs had similar affinities for IAb-3K but varied from noncrossreactive to extremely crossreactive with other peptides and MHCs. Crossreactivity correlated with a shrinking, increasingly hydrophobic TCR-ligand interface, involving fewer TCR amino acids. A few CDR1 and CDR2 amino acids dominated the most crossreactive TCR interface with MHC, including Vbeta8 48Y and 54E and Valpha4 29Y, arranged to impose the familiar diagonal orientation of TCR on MHC. These interactions contribute to MHC binding by other TCRs using related V regions, but not usually so dominantly. These data show that crossreactive TCRs can spotlight the evolutionarily conserved features of TCR-MHC interactions and that these interactions impose the diagonal docking of TCRs on MHC.
Figures

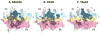




References
-
- Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. - PubMed
-
- Ashton-Rickardt PG, Bandeira A, Delaney JR, Van Kaer L, Pircher HP, Zinkernagel RM, Tonegawa S. Evidence for a differential avidity model of T cell selection in the thymus. Cell. 1994;76:651–663. - PubMed
-
- Blackman M, Yague J, Kubo R, Gay D, Coleclough C, Palmer E, Kappler J, Marrack P. The T cell repertoire may be biased in favor of MHC recognition. Cell. 1986;47:349–357. - PubMed
-
- Bluthmann H, Kisielow P, Uematsu Y, Malissen M, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. T-cell-specific deletion of T-cell receptor transgenes allows functional rearrangement of endogenous alpha- and beta-genes. Nature. 1988;334:156–159. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54 ( Pt 5):905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

