Plasmodium food vacuole plasmepsins are activated by falcipains
- PMID: 18308731
- PMCID: PMC2442342
- DOI: 10.1074/jbc.M708949200
Plasmodium food vacuole plasmepsins are activated by falcipains
Abstract
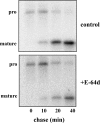
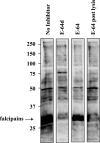
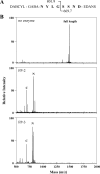
Intraerythrocytic malaria parasites use host hemoglobin as a major nutrient source. Aspartic proteases (plasmepsins) and cysteine proteases (falcipains) function in the early steps of the hemoglobin degradation pathway. There is extensive functional redundancy within and between these protease families. Plasmepsins are synthesized as integral membrane proenzymes that are activated by cleavage from the membrane. This cleavage is mediated by a maturase activity whose identity has been elusive. We have used a combination of cell biology, chemical biology, and enzymology approaches to analyze this processing event. These studies reveal that plasmepsin processing occurs primarily via the falcipains; however, if falcipain activity is blocked, autoprocessing can take place, serving as an alternate activation system. These results establish a further level of redundancy between the protease families involved in Plasmodium hemoglobin degradation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

