Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7
- PMID: 18308940
- PMCID: PMC2268811
- DOI: 10.1073/pnas.0709717105
Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7
Abstract
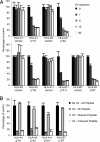
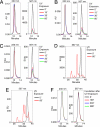
Major histocompatibility complex (MHC) class I multimer technology has become an indispensable immunological assay system to dissect antigen-specific cytotoxic CD8(+) T cell responses by flow cytometry. However, the development of high-throughput assay systems, in which T cell responses against a multitude of epitopes are analyzed, has been precluded by the fact that for each T cell epitope, a separate in vitro MHC refolding reaction is required. We have recently demonstrated that conditional ligands that disintegrate upon exposure to long-wavelength UV light can be designed for the human MHC molecule HLA-A2. To determine whether this peptide-exchange technology can be developed into a generally applicable approach for high throughput MHC based applications we set out to design conditional ligands for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Here, we describe the development and characterization of conditional ligands for this set of human MHC molecules and apply the peptide-exchange technology to identify melanoma-associated peptides that bind to HLA-A3 with high affinity. The conditional ligand technology developed here will allow high-throughput MHC-based analysis of cytotoxic T cell immunity in the vast majority of Western European individuals.
Conflict of interest statement
Conflict of interest statement: The MHC exchange technology described in this manuscript is the subject of a patent application. Based on Netherlands Cancer Institute policy on management of intellectual property, M.T., H.O. and T.N.M.S. would be entitled to a portion of received royalty income in case of future licensing.
Figures
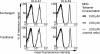

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

