Drosophila Ebi mediates Snail-dependent transcriptional repression through HDAC3-induced histone deacetylation
- PMID: 18309295
- PMCID: PMC2274935
- DOI: 10.1038/emboj.2008.26
Drosophila Ebi mediates Snail-dependent transcriptional repression through HDAC3-induced histone deacetylation
Abstract
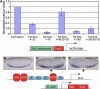
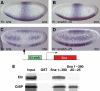
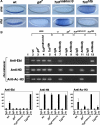
The Drosophila Snail protein is a transcriptional repressor that is necessary for mesoderm formation. Here, we identify the Ebi protein as an essential Snail co-repressor. In ebi mutant embryos, Snail target genes are derepressed in the presumptive mesoderm. Ebi and Snail interact both genetically and physically. We identify a Snail domain that is sufficient for Ebi binding, and which functions independently of another Snail co-repressor, Drosophila CtBP. This Ebi interaction domain is conserved among all insect Snail-related proteins, is a potent repression domain and is required for Snail function in transgenic embryos. In mammalian cells, the Ebi homologue TBL1 is part of the NCoR/SMRT-HDAC3 (histone deacetylase 3) co-repressor complex. We found that Ebi interacts with Drosophila HDAC3, and that HDAC3 knockdown or addition of a HDAC inhibitor impairs Snail-mediated repression in cells. In the early embryo, Ebi is recruited to a Snail target gene in a Snail-dependent manner, which coincides with histone hypoacetylation. Our results demonstrate that Snail requires the combined activities of Ebi and CtBP, and indicate that histone deacetylation is a repression mechanism in early Drosophila development.
Figures




Similar articles
-
The repressor function of snail is required for Drosophila gastrulation and is not replaceable by Escargot or Worniu.Dev Biol. 2004 May 15;269(2):411-20. doi: 10.1016/j.ydbio.2004.01.029. Dev Biol. 2004. PMID: 15110709
-
Functional interaction between the Drosophila knirps short range transcriptional repressor and RPD3 histone deacetylase.J Biol Chem. 2005 Dec 9;280(49):40757-65. doi: 10.1074/jbc.M506819200. Epub 2005 Sep 26. J Biol Chem. 2005. PMID: 16186109 Free PMC article.
-
Genetic characterization of ebi reveals its critical role in Drosophila wing growth.Fly (Austin). 2011 Oct-Dec;5(4):291-303. doi: 10.4161/fly.5.4.18276. Epub 2011 Oct 1. Fly (Austin). 2011. PMID: 22041576 Free PMC article.
-
Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor.J Biol Chem. 2005 Feb 25;280(8):7346-58. doi: 10.1074/jbc.M411675200. Epub 2004 Nov 23. J Biol Chem. 2005. PMID: 15561719
-
Insights into the function of HDAC3 and NCoR1/NCoR2 co-repressor complex in metabolic diseases.Front Mol Biosci. 2023 Aug 22;10:1190094. doi: 10.3389/fmolb.2023.1190094. eCollection 2023. Front Mol Biosci. 2023. PMID: 37674539 Free PMC article. Review.
Cited by
-
Hippo, TGF-β, and Src-MAPK pathways regulate transcription of the upd3 cytokine in Drosophila enterocytes upon bacterial infection.PLoS Genet. 2017 Nov 6;13(11):e1007091. doi: 10.1371/journal.pgen.1007091. eCollection 2017 Nov. PLoS Genet. 2017. PMID: 29108021 Free PMC article.
-
Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family.Genes Dev. 2015 Jan 1;29(1):48-62. doi: 10.1101/gad.252122.114. Genes Dev. 2015. PMID: 25561495 Free PMC article.
-
HDAC3 selectively represses CREB3-mediated transcription and migration of metastatic breast cancer cells.Cell Mol Life Sci. 2010 Oct;67(20):3499-510. doi: 10.1007/s00018-010-0388-5. Epub 2010 May 15. Cell Mol Life Sci. 2010. PMID: 20473547 Free PMC article.
-
Targeting histone deacetylase-3 blocked epithelial-mesenchymal plasticity and metastatic dissemination in gastric cancer.Cell Biol Toxicol. 2023 Oct;39(5):1873-1896. doi: 10.1007/s10565-021-09673-2. Epub 2022 Jan 1. Cell Biol Toxicol. 2023. PMID: 34973135 Free PMC article.
-
Epigenetic inheritance and gene expression regulation in early Drosophila embryos.EMBO Rep. 2024 Oct;25(10):4131-4152. doi: 10.1038/s44319-024-00245-z. Epub 2024 Sep 16. EMBO Rep. 2024. PMID: 39285248 Free PMC article. Review.
References
-
- Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M (1991) The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development 111: 983–992 - PubMed
-
- Barrallo-Gimeno A, Nieto MA (2005) The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132: 3151–3161 - PubMed
-
- Chinnadurai G (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell 9: 213–224 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

