Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52
- PMID: 18310075
- PMCID: PMC2335352
- DOI: 10.1074/jbc.M800763200
Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52
Abstract
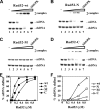
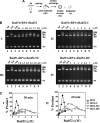
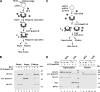
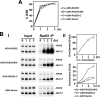
A helical filament of Rad51 on single-strand DNA (ssDNA), called the presynaptic filament, catalyzes DNA joint formation during homologous recombination. Rad52 facilitates presynaptic filament assembly, and this recombination mediator activity is thought to rely on the interactions of Rad52 with Rad51, the ssDNA-binding protein RPA, and ssDNA. The N-terminal region of Rad52, which has DNA binding activity and an oligomeric structure, is thought to be crucial for mediator activity and recombination. Unexpectedly, we find that the C-terminal region of Rad52 also harbors a DNA binding function. Importantly, the Rad52 C-terminal portion alone can promote Rad51 presynaptic filament assembly. The middle portion of Rad52 associates with DNA-bound RPA and contributes to the recombination mediator activity. Accordingly, expression of a protein species that harbors the middle and C-terminal regions of Rad52 in the rad52 Delta327 background enhances the association of Rad51 protein with a HO-made DNA double-strand break and partially complements the methylmethane sulfonate sensitivity of the mutant cells. Our results provide a mechanistic framework for rationalizing the multi-faceted role of Rad52 in recombination and DNA repair.
Figures




References
-
- Cox, M. M., Goodman, M. F., Kreuzer, K. N., Sherratt, D. J., Sandler, S. J., and Marians, K. J. (2000) Nature 404 37–41 - PubMed
-
- Heller, R. C., and Marians, K. J. (2006) Nat. Rev. Mol. Cell Biol. 7 932–943 - PubMed
-
- Jasin, M. (2002) Oncogene 21 8981–8993 - PubMed
-
- D'Andrea, A. D. (2003) Genes Dev. 17 1933–1936 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

