GroEL stimulates protein folding through forced unfolding
- PMID: 18311152
- PMCID: PMC3744391
- DOI: 10.1038/nsmb.1394
GroEL stimulates protein folding through forced unfolding
Abstract
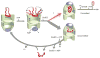
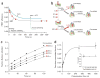
Many proteins cannot fold without the assistance of chaperonin machines like GroEL and GroES. The nature of this assistance, however, remains poorly understood. Here we demonstrate that unfolding of a substrate protein by GroEL enhances protein folding. We first show that capture of a protein on the open ring of a GroEL-ADP-GroES complex, GroEL's physiological acceptor state for non-native proteins in vivo, leaves the substrate protein in an unexpectedly compact state. Subsequent binding of ATP to the same GroEL ring causes rapid, forced unfolding of the substrate protein. Notably, the fraction of the substrate protein that commits to the native state following GroES binding and protein release into the GroEL-GroES cavity is proportional to the extent of substrate-protein unfolding. Forced protein unfolding is thus a central component of the multilayered stimulatory mechanism used by GroEL to drive protein folding.
Figures
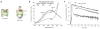
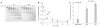
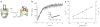
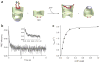

References
-
- Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. - PubMed
-
- Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147–154. - PubMed
-
- Grantcharova V, Alm EJ, Baker D, Horwich AL. Mechanisms of protein folding. Curr Opin Struct Biol. 2001;11:70–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

