The CBS subdomain of inosine 5'-monophosphate dehydrogenase regulates purine nucleotide turnover
- PMID: 18312263
- PMCID: PMC2279236
- DOI: 10.1111/j.1365-2958.2008.06153.x
The CBS subdomain of inosine 5'-monophosphate dehydrogenase regulates purine nucleotide turnover
Abstract
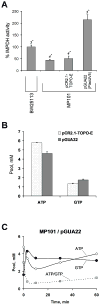
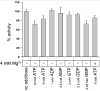
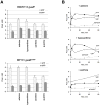
Inosine 5'-monophosphate dehydrogenase (IMPDH) catalyses the rate-limiting step in guanine nucleotide biosynthesis. IMPDH has an evolutionary conserved CBS subdomain of unknown function. The subdomain can be deleted without impairing the in vitro IMPDH catalytic activity and is the site for mutations associated with human retinitis pigmentosa. A guanine-prototrophic Escherichia coli strain, MP101, was constructed with the subdomain sequence deleted from the chromosomal gene for IMPDH. The ATP content was substantially elevated in MP101 whereas the GTP content was slighty reduced. The activities of IMPDH, adenylosuccinate synthetase and GMP reductase were two to threefold lower in MP101 crude extracts compared with the BW25113 wild-type strain. Guanine induced a threefold reduction in the MP101 ATP pool and a fourfold increase in the GTP pool within 10 min of addition to growing cells; this response does not result from the reduced IMPDH activity or starvation for guanylates. In vivo kinetic analysis using 14-C tracers and 33-P pulse-chasing revealed mutation-associated changes in purine nucleotide fluxes and turnover rates. We conclude that the CBS subdomain of IMPDH may coordinate the activities of the enzymes of purine nucleotide metabolism and is essential for maintaining the normal ATP and GTP pool sizes in E. coli.
Figures




References
-
- Ataullakhanov FI, Vitvitsky VM. What determines the intracellular ATP concentration. Biosci Rep. 2002;22:501–511. - PubMed
-
- Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. - PubMed
-
- Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

