A heteroduplex assay for the rapid detection of dual Human Immunodeficiency Virus Type 1 infections
- PMID: 18314205
- PMCID: PMC3773955
- DOI: 10.1016/j.jviromet.2008.01.015
A heteroduplex assay for the rapid detection of dual Human Immunodeficiency Virus Type 1 infections
Abstract
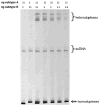
The predominance of circulating and unique recombinant forms (URFs) of Human Immunodeficiency Virus Type 1 (HIV-1) in Cameroon suggests that dual infection occurs frequently in this region. Despite the potential impact of these infections on the evolution of HIV diversity, relatively few have been detected. The failure to detect dual infections may be attributable to the laborious and costly sequence analysis involved in their identification. As such, there is a need for a cost-effective, more rapid method to efficiently distinguish this subset of HIV-positive individuals, particularly in regions where HIV diversity is broad. In the present study, the heteroduplex assay (HDA) was developed to detect dual HIV-1 infection. This assay was validated on sequential specimens obtained from 20 HIV+ study subjects, whose single or dual infection status was determined by standard sequence analysis. By mixing gag fragments amplified from the sequential specimens from each study subject in HDA reactions, it was shown that single and dual infection status correlated with the absence and presence, respectively, of heteroduplex bands upon gel electrophoresis. Therefore, this novel assay is capable of identifying dual infections with a sensitivity and specificity equivalent to that of sequence analysis. Given the impact of dual infection on viral recombination and diversity, this simple technique will be beneficial to understanding HIV-1 evolution within an individual, as well as at a population level, in West-Central Africa and globally.
Figures


References
-
- Delwart EL, Shpaer EG, Louwagie J, McCutchan FE, Grez M, Rubsamen-Waigmann H, Mullins JI. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262 (5137):1257–1261. - PubMed
-
- Fang G, Zhu G, Burger H, Keithly JS, Weiser B. Minimizing DNA recombination during long RT-PCR. J Virol Methods. 1998;76 (1/2):139–148. - PubMed
-
- Felsenstein J. Confidence limits on Phylogenies: an approach using the bootstrap. Evolution. 1985;39 (4):783–791. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

