Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme
- PMID: 18314503
- PMCID: PMC2271362
- DOI: 10.1261/rna.824308
Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme
Abstract
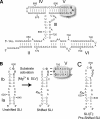
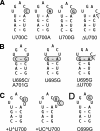
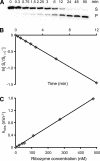
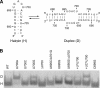
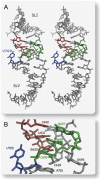
Substrate recognition by the VS ribozyme involves a magnesium-dependent loop/loop interaction between the SLI substrate and the SLV hairpin from the catalytic domain. Recent NMR studies of SLV demonstrated that magnesium ions stabilize a U-turn loop structure and trigger a conformational change for the extruded loop residue U700, suggesting a role for U700 in SLI recognition. Here, we kinetically characterized VS ribozyme mutants to evaluate the contribution of U700 and other SLV loop residues to SLI recognition. To help interpret the kinetic data, we structurally characterized the SLV mutants by NMR spectroscopy and generated a three-dimensional model of the SLI/SLV complex by homology modeling with MC-Sym. We demonstrated that the mutation of U700 by A, C, or G does not significantly affect ribozyme activity, whereas deletion of U700 dramatically impairs this activity. The U700 backbone is likely important for SLI recognition, but does not appear to be required for either the structural integrity of the SLV loop or for direct interactions with SLI. Thus, deletion of U700 may affect other aspects of SLI recognition, such as magnesium ion binding and SLV loop dynamics. As part of our NMR studies, we developed a convenient assay based on detection of unusual (31)P and (15)N N7 chemical shifts to probe the formation of U-turn structures in RNAs. Our model of the SLI/SLV complex, which is compatible with biochemical data, leads us to propose novel interactions at the loop I/loop V interface.
Figures

Similar articles
-
A remarkably stable kissing-loop interaction defines substrate recognition by the Neurospora Varkud Satellite ribozyme.RNA. 2014 Sep;20(9):1451-64. doi: 10.1261/rna.046144.114. Epub 2014 Jul 22. RNA. 2014. PMID: 25051972 Free PMC article.
-
Structural insights into substrate recognition by the Neurospora Varkud satellite ribozyme: importance of U-turns at the kissing-loop junction.Biochemistry. 2014 Jan 14;53(1):258-69. doi: 10.1021/bi401491g. Epub 2013 Dec 17. Biochemistry. 2014. PMID: 24325625 Free PMC article.
-
Mg2+ Binding Promotes SLV as a Scaffold in Varkud Satellite Ribozyme SLI-SLV Kissing Loop Junction.Biophys J. 2017 Jul 25;113(2):313-320. doi: 10.1016/j.bpj.2017.06.008. Epub 2017 Jun 29. Biophys J. 2017. PMID: 28669407 Free PMC article.
-
The Varkud satellite ribozyme.RNA. 2004 Feb;10(2):151-8. doi: 10.1261/rna.5217104. RNA. 2004. PMID: 14730013 Free PMC article. Review.
-
Insights into RNA structure and dynamics from recent NMR and X-ray studies of the Neurospora Varkud satellite ribozyme.Wiley Interdiscip Rev RNA. 2017 Sep;8(5):e1421. doi: 10.1002/wrna.1421. Epub 2017 Apr 6. Wiley Interdiscip Rev RNA. 2017. PMID: 28382748 Free PMC article. Review.
Cited by
-
Stepwise assembly of multiple Lin28 proteins on the terminal loop of let-7 miRNA precursors.Nucleic Acids Res. 2014 Apr;42(7):4615-28. doi: 10.1093/nar/gkt1391. Epub 2014 Jan 21. Nucleic Acids Res. 2014. PMID: 24452802 Free PMC article.
-
Stem-Loop V of Varkud Satellite RNA Exhibits Characteristics of the Mg(2+) Bound Structure in the Presence of Monovalent Ions.J Phys Chem B. 2015 Sep 24;119(38):12355-64. doi: 10.1021/acs.jpcb.5b05190. Epub 2015 Sep 14. J Phys Chem B. 2015. PMID: 26328924 Free PMC article.
-
Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28.Nucleic Acids Res. 2012 Feb;40(4):1767-77. doi: 10.1093/nar/gkr808. Epub 2011 Oct 19. Nucleic Acids Res. 2012. PMID: 22013165 Free PMC article.
-
Formation of an active site in trans by interaction of two complete Varkud Satellite ribozymes.RNA. 2009 Oct;15(10):1822-6. doi: 10.1261/rna.1759009. Epub 2009 Aug 24. RNA. 2009. PMID: 19703941 Free PMC article.
-
Rational engineering of the Neurospora VS ribozyme to allow substrate recognition via different kissing-loop interactions.Nucleic Acids Res. 2016 Aug 19;44(14):6924-34. doi: 10.1093/nar/gkw401. Epub 2016 May 10. Nucleic Acids Res. 2016. PMID: 27166370 Free PMC article.
References
-
- Andersen, A., Collins, R.A. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell. 2000;5:469–478. - PubMed
-
- Beattie, T.L., Collins, R.A. Identification of functional domains in the self-cleaving Neurospora VS ribozyme using damage selection. J. Mol. Biol. 1997;267:830–840. - PubMed
-
- Cabello-Villegas, J., Tworowska, I., Nikonowicz, E.P. Metal ion stabilization of the U-turn of the A37 N6-dimethylallyl-modified anticodon stem–loop of Escherichia coli tRNAPhe . Biochemistry. 2004;43:55–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
