Population diversity and dynamics of Streptococcus mitis, Streptococcus oralis, and Streptococcus infantis in the upper respiratory tracts of adults, determined by a nonculture strategy
- PMID: 18316382
- PMCID: PMC2346710
- DOI: 10.1128/IAI.01511-07
Population diversity and dynamics of Streptococcus mitis, Streptococcus oralis, and Streptococcus infantis in the upper respiratory tracts of adults, determined by a nonculture strategy
Abstract
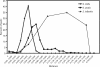
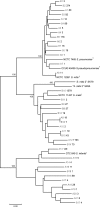
We reinvestigated the clonal diversity and dynamics of Streptococcus mitis and two other abundant members of the commensal microbiota of the upper respiratory tract, Streptococcus oralis and Streptococcus infantis, to obtain information about the origin of frequently emerging clones in this habitat. A culture-independent method was used, based on cloning and sequencing of PCR amplicons of the housekeeping gene gdh, which shows remarkable, yet species-specific, genetic polymorphism. Samples were collected from all potential ecological niches in the oral cavity and pharynx of two adults on two occasions separated by 2 years. Based on analysis of close to 10,000 sequences, significant diversity was observed in populations of all three species. Fluctuations in the relative proportions of individual clones and species were observed over time. While a few clones dominated, the proportions of most clones were very small. The results show that the frequent turnover of S. mitis, S. oralis, and S. infantis clones observed by cultivation can be explained by fluctuations in the relative proportions of clones, most of which are below the level of detection by the traditional culture technique, possibly combined with loss and acquisition from contacts. These findings provide a platform for understanding the mechanisms that govern the balance within the complex microbiota at mucosal sites and between the microbiota and the mucosal immune system of the host.
Figures


References
-
- Arbique, J. C., C. Poyart, P. Trieu-Cuot, G. Quesne, M. G. Carvalho, A. G. Steigerwalt, R. E. Morey, D. Jackson, R. J. Davidson, and R. R. Facklam. 2004. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 424686-4696. - PMC - PubMed
-
- Bowden, G. H., J. Ekstrand, B. McNaughton, and S. J. Challacombe. 1990. Association of selected bacteria with the lesions of root surface caries. Oral Microbiol. Immunol. 5346-351. - PubMed
-
- Carratala, J., B. Roson, A. Fernandez-Sevilla, F. Alcaide, and F. Gudiol. 1998. Bacteremic pneumonia in neutropenic patients with cancer: causes, empirical antibiotic therapy, and outcome. Arch. Intern. Med. 158868-872. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

