A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia
- PMID: 18316384
- PMCID: PMC2346693
- DOI: 10.1128/IAI.01338-07
A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia
Abstract
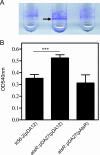
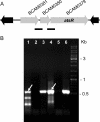
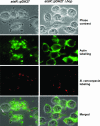
Burkholderia cenocepacia is an important opportunistic pathogen causing serious chronic infections in patients with cystic fibrosis (CF). Adaptation of B. cenocepacia to the CF airways may play an important role in the persistence of the infection. We have identified a sensor kinase-response regulator (BCAM0379) named AtsR in B. cenocepacia K56-2 that shares 19% amino acid identity with RetS from Pseudomonas aeruginosa. atsR inactivation led to increased biofilm production and a hyperadherent phenotype in both abiotic surfaces and lung epithelial cells. Also, the atsR mutant overexpressed and hypersecreted an Hcp-like protein known to be specifically secreted by the type VI secretion system (T6SS) in other gram-negative bacteria. Amoeba plaque assays demonstrated that the atsR mutant was more resistant to Dictyostelium predation than the wild-type strain and that this phenomenon was T6SS dependent. Macrophage infection assays also demonstrated that the atsR mutant induces the formation of actin-mediated protrusions from macrophages that require a functional Hcp-like protein, suggesting that the T6SS is involved in actin rearrangements. Three B. cenocepacia transposon mutants that were found in a previous study to be impaired for survival in chronic lung infection model were mapped to the T6SS gene cluster, indicating that the T6SS is required for infection in vivo. Together, our data show that AtsR is involved in the regulation of genes required for virulence in B. cenocepacia K56-2, including genes encoding a T6SS.
Figures




References
-
- Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5719-729. - PubMed
-
- Corbett, C. R., M. N. Burtnick, C. Kooi, D. E. Woods, and P. A. Sokol. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 1492263-2271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

