An antibiotic-responsive mouse model of fulminant ulcerative colitis
- PMID: 18318596
- PMCID: PMC2270287
- DOI: 10.1371/journal.pmed.0050041
An antibiotic-responsive mouse model of fulminant ulcerative colitis
Abstract
Background: The constellation of human inflammatory bowel disease (IBD) includes ulcerative colitis and Crohn's disease, which both display a wide spectrum in the severity of pathology. One theory is that multiple genetic hits to the host immune system may contribute to the susceptibility and severity of IBD. However, experimental proof of this concept is still lacking. Several genetic mouse models that each recapitulate some aspects of human IBD have utilized a single gene defect to induce colitis. However, none have produced pathology clearly distinguishable as either ulcerative colitis or Crohn's disease, in part because none of them reproduce the most severe forms of disease that are observed in human patients. This lack of severe IBD models has posed a challenge for research into pathogenic mechanisms and development of new treatments. We hypothesized that multiple genetic hits to the regulatory machinery that normally inhibits immune activation in the intestine would generate more severe, reproducible pathology that would mimic either ulcerative colitis or Crohn's disease.
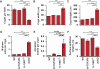
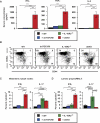
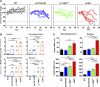
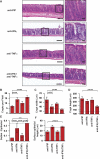
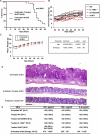
Methods and findings: We generated a novel mouse line (dnKO) that possessed defects in both TGFbetaRII and IL-10R2 signaling. These mice rapidly and reproducibly developed a disease resembling fulminant human ulcerative colitis that was quite distinct from the much longer and more variable course of pathology observed previously in mice possessing only single defects. Pathogenesis was driven by uncontrolled production of proinflammatory cytokines resulting in large part from T cell activation. The disease process could be significantly ameliorated by administration of antibodies against IFNgamma and TNFalpha and was completely inhibited by a combination of broad-spectrum antibiotics.
Conclusions: Here, we develop to our knowledge the first mouse model of fulminant ulcerative colitis by combining multiple genetic hits in immune regulation and demonstrate that the resulting disease is sensitive to both anticytokine therapy and broad-spectrum antibiotics. These findings indicated the IL-10 and TGFbeta pathways synergize to inhibit microbially induced production of proinflammatory cytokines, including IFNgamma and TNFalpha, which are known to play a role in the pathogenesis of human ulcerative colitis. Our findings also provide evidence that broad-spectrum antibiotics may have an application in the treatment of patients with ulcerative colitis. This model system will be useful in the future to explore the microbial factors that induce immune activation and characterize how these interactions produce disease.
Conflict of interest statement
Figures



Comment in
-
You see UC: an animal model of ulcerative colitis.Gastroenterology. 2008 Dec;135(6):2149-50. doi: 10.1053/j.gastro.2008.10.062. Epub 2008 Nov 8. Gastroenterology. 2008. PMID: 19000681 No abstract available.
References
-
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. - PubMed
-
- Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, et al. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. - PubMed
-
- Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, et al. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. - PubMed
-
- Rudolph U, Finegold MJ, Rich SS, Harriman GR, Srinivasan Y, et al. Ulcerative colitis and adenocarcinoma of the colon in G alpha i2-deficient mice. Nat Genet. 1995;10:143–150. - PubMed
-
- Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360:225–231. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

