Normalization of oligonucleotide arrays based on the least-variant set of genes
- PMID: 18318917
- PMCID: PMC2324100
- DOI: 10.1186/1471-2105-9-140
Normalization of oligonucleotide arrays based on the least-variant set of genes
Abstract
Background: It is well known that the normalization step of microarray data makes a difference in the downstream analysis. All normalization methods rely on certain assumptions, so differences in results can be traced to different sensitivities to violation of the assumptions. Illustrating the lack of robustness, in a striking spike-in experiment all existing normalization methods fail because of an imbalance between up- and down-regulated genes. This means it is still important to develop a normalization method that is robust against violation of the standard assumptions
Results: We develop a new algorithm based on identification of the least-variant set (LVS) of genes across the arrays. The array-to-array variation is evaluated in the robust linear model fit of pre-normalized probe-level data. The genes are then used as a reference set for a non-linear normalization. The method is applicable to any existing expression summaries, such as MAS5 or RMA.
Conclusion: We show that LVS normalization outperforms other normalization methods when the standard assumptions are not satisfied. In the complex spike-in study, LVS performs similarly to the ideal (in practice unknown) housekeeping-gene normalization. An R package called lvs is available in http://www.meb.ki.se/~yudpaw.
Figures
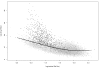
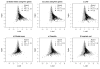

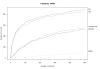

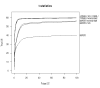
References
-
- Hartemink A, Gifford D, Jaakkola T, Young R. Maximum likelihood estimation of optimal scaling factors for expression array normalizations. IN SPIE Bios. 2001.
-
- Affymetrix Statistical Algorithms Description Document. 2002.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

