Epigenomic profiling reveals DNA-methylation changes associated with major psychosis
- PMID: 18319075
- PMCID: PMC2427301
- DOI: 10.1016/j.ajhg.2008.01.008
Epigenomic profiling reveals DNA-methylation changes associated with major psychosis
Abstract
Epigenetic misregulation is consistent with various non-Mendelian features of schizophrenia and bipolar disorder. To date, however, few studies have investigated the role of DNA methylation in major psychosis, and none have taken a genome-wide epigenomic approach. In this study we used CpG-island microarrays to identify DNA-methylation changes in the frontal cortex and germline associated with schizophrenia and bipolar disorder. In the frontal cortex we find evidence for psychosis-associated DNA-methylation differences in numerous loci, including several involved in glutamatergic and GABAergic neurotransmission, brain development, and other processes functionally linked to disease etiology. DNA-methylation changes in a significant proportion of these loci correspond to reported changes of steady-state mRNA level associated with psychosis. Gene-ontology analysis highlighted epigenetic disruption to loci involved in mitochondrial function, brain development, and stress response. Methylome network analysis uncovered decreased epigenetic modularity in both the brain and the germline of affected individuals, suggesting that systemic epigenetic dysfunction may be associated with major psychosis. We also report evidence for a strong correlation between DNA methylation in the MEK1 gene promoter region and lifetime antipsychotic use in schizophrenia patients. Finally, we observe that frontal-cortex DNA methylation in the BDNF gene is correlated with genotype at a nearby nonsynonymous SNP that has been previously associated with major psychosis. Our data are consistent with the epigenetic theory of major psychosis and suggest that DNA-methylation changes are important to the etiology of schizophrenia and bipolar disorder.
Figures
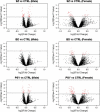



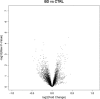
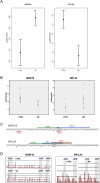
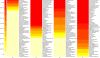

References
-
- Petronis A. The origin of schizophrenia: genetic thesis, epigenetic antithesis, and resolving synthesis. Biol Psychiatry. 2004;55:965–970. - PubMed
-
- McGuffin P., Asherson P., Owen M., Farmer A. The strength of the genetic effect. Is there room for an environmental influence in the aetiology of schizophrenia? Br. J. Psychiatry. 1994;164:593–599. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

