Autophagy-physiology and pathophysiology
- PMID: 18320203
- PMCID: PMC2668654
- DOI: 10.1007/s00418-008-0406-y
Autophagy-physiology and pathophysiology
Abstract
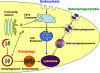
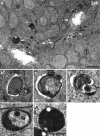
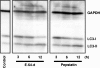
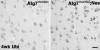
"Autophagy" is a highly conserved pathway for degradation, by which wasted intracellular macromolecules are delivered to lysosomes, where they are degraded into biologically active monomers such as amino acids that are subsequently re-used to maintain cellular metabolic turnover and homeostasis. Recent genetic studies have shown that mice lacking an autophagy-related gene (Atg5 or Atg7) cannot survive longer than 12 h after birth because of nutrient shortage. Moreover, tissue-specific impairment of autophagy in central nervous system tissue causes massive loss of neurons, resulting in neurodegeneration, while impaired autophagy in liver tissue causes accumulation of wasted organelles, leading to hepatomegaly. Although autophagy generally prevents cell death, our recent study using conditional Atg7-deficient mice in CNS tissue has demonstrated the presence of autophagic neuron death in the hippocampus after neonatal hypoxic/ischemic brain injury. Thus, recent genetic studies have shown that autophagy is involved in various cellular functions. In this review, we introduce physiological and pathophysiological roles of autophagy.
Figures






References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.2353/ajpath.2006.051066', 'is_inner': False, 'url': 'https://doi.org/10.2353/ajpath.2006.051066'}, {'type': 'PMC', 'value': 'PMC1780162', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC1780162/'}, {'type': 'PubMed', 'value': '16877357', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/16877357/'}]}
- Adhami F, Liao G, Morozov YM, Schloemer A, Schmithorst VJ, Lorenz JN, Dunn RS, Vorhees CV, Wills-Karp M, Degen JL, Davis RJ, Mizushima N, Rakic P, Dardzinski BJ, Holland SK, Sharp FR, Kuan CY (2006) Cerebral ischemia-hypoxia induces intravascular coagulation and autophagy. Am J Pathol 169:566–583 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '17035724', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17035724/'}]}
- Adhami F, Schloemer A, Kuan CY (2007) The roles of autophagy in cerebral ischemia. Autophagy 3:42–44 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/j.febslet.2004.12.058', 'is_inner': False, 'url': 'https://doi.org/10.1016/j.febslet.2004.12.058'}, {'type': 'PubMed', 'value': '15670810', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/15670810/'}]}
- Ardley HC, Hung CC, Robinson PA (2005) The aggravating role of the ubiquitin–proteasome system in neurodegeneration. FEBS Lett 579:571–576 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PMC', 'value': 'PMC2013521', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC2013521/'}, {'type': 'PubMed', 'value': '4300890', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/4300890/'}]}
- Arstila AU, Trump BF (1968) Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol 53:687–733 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1083/jcb.12.1.198', 'is_inner': False, 'url': 'https://doi.org/10.1083/jcb.12.1.198'}, {'type': 'PMC', 'value': 'PMC2106008', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC2106008/'}, {'type': 'PubMed', 'value': '13862833', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/13862833/'}]}
- Ashford TP, Porter KR (1962) Cytoplasmic components in hepatic cell lysosomes. J Cell Biol 12:198–202 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

