Predicting proteolytic sites in extracellular proteins: only halfway there
- PMID: 18321887
- PMCID: PMC7109841
- DOI: 10.1093/bioinformatics/btn084
Predicting proteolytic sites in extracellular proteins: only halfway there
Abstract
Motivation: Many secretory proteins are synthesized as inactive precursors that must undergo post-translational proteolysis in order to mature and become active. In the current study, we address the challenge of sequence-based discovery of proteolytic sites in secreted proteins using machine learning.
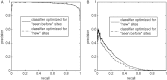
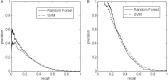
Results: The results revealed that only half of the extracellular proteolytic sites are currently annotated, leaving over 3600 unannotated ones. Furthermore, we have found that only 6% of the unannotated sites are similar to known proteolytic sites, whereas the remaining 94% do not share significant similarity with any annotated proteolytic site. The computational challenges in these two cases are very different. While the precision in detecting the former group is close to perfect, only a mere 22% of the latter group were detected with a precision of 80%. The applicability of the classifier is demonstrated through members of the FGF family, in which we verified the conservation of physiologically-relevant proteolytic sites in homologous proteins.
Figures


References
-
- Anderson NL, et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol. Cell Proteomics. 2004;3:311–326. - PubMed
-
- Antoine M, et al. NH2-terminal cleavage of xenopus fibroblast growth factor 3 is necessary for optimal biological activity and receptor binding. Cell Growth Differ. 2000;11:593–605. - PubMed
-
- Basak A. Inhibitors of proprotein convertases. J. Mol. Med. 2005;83:844–855. - PubMed
-
- Bendtsen JD, et al. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. - PubMed

