Human RNA "rumor" viruses: the search for novel human retroviruses in chronic disease
- PMID: 18322038
- PMCID: PMC2268285
- DOI: 10.1128/MMBR.00033-07
Human RNA "rumor" viruses: the search for novel human retroviruses in chronic disease
Abstract
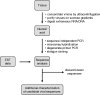
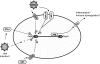
Retroviruses are an important group of pathogens that cause a variety of diseases in humans and animals. Four human retroviruses are currently known, including human immunodeficiency virus type 1, which causes AIDS, and human T-lymphotropic virus type 1, which causes cancer and inflammatory disease. For many years, there have been sporadic reports of additional human retroviral infections, particularly in cancer and other chronic diseases. Unfortunately, many of these putative viruses remain unproven and controversial, and some retrovirologists have dismissed them as merely "human rumor viruses." Work in this field was last reviewed in depth in 1984, and since then, the molecular techniques available for identifying and characterizing retroviruses have improved enormously in sensitivity. The advent of PCR in particular has dramatically enhanced our ability to detect novel viral sequences in human tissues. However, DNA amplification techniques have also increased the potential for false-positive detection due to contamination. In addition, the presence of many families of human endogenous retroviruses (HERVs) within our DNA can obstruct attempts to identify and validate novel human retroviruses. Here, we aim to bring together the data on "novel" retroviral infections in humans by critically examining the evidence for those putative viruses that have been linked with disease and the likelihood that they represent genuine human infections. We provide a background to the field and a discussion of potential confounding factors along with some technical guidelines. In addition, some of the difficulties associated with obtaining formal proof of causation for common or ubiquitous agents such as HERVs are discussed.
Figures




Comment in
-
What is the epidemiology of human mammary tumor virus?Int J Gynecol Cancer. 2014 Mar;24(3):382. doi: 10.1097/IGC.0000000000000071. Int J Gynecol Cancer. 2014. PMID: 24557431 No abstract available.
References
-
- Acha-Orbea, H., D. Finke, A. Attinger, S. Schmid, N. Wehrli, S. Vacheron, I. Xenarios, L. Scarpellino, K. M. Toellner, I. C. MacLennan, and S. A. Luther. 1999. Interplays between mouse mammary tumor virus and the cellular and humoral immune response. Immunol. Rev. 168287-303. - PubMed
-
- Acha-Orbea, H., and H. R. MacDonald. 1995. Superantigens of mouse mammary tumor virus. Annu. Rev. Immunol. 13459-486. - PubMed
-
- Achong, B. G., and M. A. Epstein. 1978. Preliminary seroepidemiological studies on the human syncytial virus. J. Gen. Virol. 40175-181. - PubMed
-
- Achong, B. G., P. W. Mansell, M. A. Epstein, and P. Clifford. 1971. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 46299-307. - PubMed
-
- Achong, B. G., P. A. Trumper, and B. C. Giovanella. 1976. C-type virus particles in human tumours transplanted into nude mice. Br. J. Cancer 34203-206. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

