Moonlighting proteins in yeasts
- PMID: 18322039
- PMCID: PMC2268286
- DOI: 10.1128/MMBR.00036-07
Moonlighting proteins in yeasts
Abstract
Proteins able to participate in unrelated biological processes have been grouped under the generic name of moonlighting proteins. Work with different yeast species has uncovered a great number of moonlighting proteins and shown their importance for adequate functioning of the yeast cell. Moonlighting activities in yeasts include such diverse functions as control of gene expression, organelle assembly, and modification of the activity of metabolic pathways. In this review, we consider several well-studied moonlighting proteins in different yeast species, paying attention to the experimental approaches used to identify them and the evidence that supports their participation in the unexpected function. Usually, moonlighting activities have been uncovered unexpectedly, and up to now, no satisfactory way to predict moonlighting activities has been found. Among the well-characterized moonlighting proteins in yeasts, enzymes from the glycolytic pathway appear to be prominent. For some cases, it is shown that despite close phylogenetic relationships, moonlighting activities are not necessarily conserved among yeast species. Organisms may utilize moonlighting to add a new layer of regulation to conventional regulatory networks. The existence of this type of proteins in yeasts should be taken into account when designing mutant screens or in attempts to model or modify yeast metabolism.
Figures
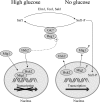
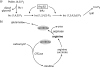
References
-
- Ahuatzi, D., P. Herrero, T. de la Cera, and F. Moreno. 2004. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 27914440-14446. - PubMed
-
- Ahuatzi, D., A. Riera, R. Pelaez, P. Herrero, and F. Moreno. 2007. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 2824485-4493. - PubMed
-
- Aigle, M., and F. Lacroute. 1975. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet. 136327-335. - PubMed
-
- Anders, A., H. Lilie, K. Franke, L. Kapp, J. Stelling, E. D. Gilles, and K. D. Breunig. 2006. The galactose switch in Kluyveromyces lactis depends on nuclear competition between Gal4 and Gal1 for Gal80 binding. J. Biol. Chem. 28129337-29348. - PubMed
-
- Armstrong, R. N. 1997. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 102-18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

