Early neuronal and glial fate restriction of embryonic neural stem cells
- PMID: 18322099
- PMCID: PMC6671176
- DOI: 10.1523/JNEUROSCI.5497-07.2008
Early neuronal and glial fate restriction of embryonic neural stem cells
Abstract
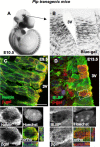
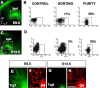
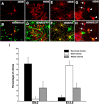
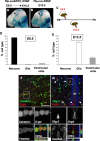
The question of how neurons and glial cells are generated during the development of the CNS has over time led to two alternative models: either neuroepithelial cells are capable of giving rise to neurons first and to glial cells at a later stage (switching model), or they are intrinsically committed to generate one or the other (segregating model). Using the developing diencephalon as a model and by selecting a subpopulation of ventricular cells, we analyzed both in vitro, using clonal analysis, and in vivo, using inducible Cre/loxP fate mapping, the fate of neuroepithelial and radial glial cells generated at different time points during embryonic development. We found that, during neurogenic periods [embryonic day 9.5 (E9.5) to 12.5], proteolipid protein (plp)-expressing cells were lineage-restricted neuronal precursors, but later in embryogenesis, during gliogenic periods (E13.5 to early postnatal), plp-expressing cells were lineage-restricted glial precursors. In addition, we show that glial cells forming at E13.5 arise from a new pool of neuroepithelial progenitors distinct from neuronal progenitors cells, which lends support to the segregating model.
Figures


References
-
- Anderson DJ. Stem cells and pattern formation in the nervous system: the possible versus the actual. Neuron. 2001;30:19–35. - PubMed
-
- Battiste J, Helms AW, Kim EJ, Savage TK, Lagace DC, Mandyam CD, Eisch AJ, Miyoshi G, Johnson JE. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development. 2007;134:285–293. - PubMed
-
- Bellefroid EJ, Bourguignon C, Hollemann T, Ma Q, Anderson DJ, Kintner C, Pieler T. X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell. 1996;87:1191–1202. - PubMed
-
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. - PubMed
-
- Bhat RV, Axt KJ, Fosnaugh JS, Smith KJ, Johnson KA, Hill DE, Kinzler KW, Baraban JM. Expression of the APC tumor suppressor protein in oligodendroglia. Glia. 1996;17:169–174. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
